This blog post looks at the segments you got from your ancestors. It will be an effort to outline what you should expect from various ancestors – to give you an overview of how you got your segments, and how they are arranged. Your DNA is like a big jigsaw puzzle, with many different and unique pieces – this will let you see a picture that might help you solve the puzzle. Of course, your picture will be somewhat different. DNA is very random, and there is wide variation in what you actually got. Nevertheless, there are averages, and there are some rules. I hope this post will give you an understanding of the big picture, as well as some detail, and help you work with atDNA segments.
There are two ways to look at your DNA: Top-Down and Bottom-Up.
Top-Down is the way you got your DNA segments – from your distant ancestors (from the top of your Tree), down to you (at the bottom of your Tree). This Top-Down explanation often includes many ancestors and DNA segments (lots of colors in the diagrams), and can get quite complex after a very few generations. I’ll attempt a simple version of the Top-Down look in a separate blog post.
Bottom-Up tends to be the way we look at our DNA – we start with all of our own DNA and divide it into maternal and paternal sides; and then determine the segments from grandparents and Great grandparents, etc. We work from ourselves (at the bottom of our Tree), up the Tree as far as we can go. This is the “look” that will be described below.
Before we start, there are three points to make:
- We are talking about ancestral segments (see What is a segment). These are all segments you got from your ancestors.
- We are not talking about shared segments with Matches. This discussion is only about you and your ancestors and the segments from them. There will be more about shared segments in later other blog posts.
- DNA is random. This is a rough model; a general picture of our DNA segments. Please don’t get lost in the technical details or in an unusual situation. This is all about what you are likely to encounter – it’s definitely not a one-size-fits all description. Your DNA is different, but the general principles below will apply.
Before we discuss shared segments (in a later blog post), it’s important first to understand how our DNA is made up of segments from different ancestors, and generally what to expect about these segments – how they are arranged in each chromosome. This understanding of ancestral segments will help you understand shared segments later.
Three ground rules:
- This discussion will be about autosomal DNA (atDNA) – the numbered chromosomes (chromosome 1 to 22)
- This discussion will focus on one parent – Mother. The concepts apply equally to either parent.
- Examples will usually use chromosome 5.
So let’s get started…
Parents
You get many large segments from your Mother. They are exactly the size of each chromosome – because each segment is a chromosome. Your Mother gave you one of each of the 22 autosomes (chromosomes 1 to 22). In each case this is a large segment from the beginning of a chromosome (the first base pair) to the end of each chromosome (the last base pair). You probably already knew that you got one set of chromosomes from your Mother, but you may not have thought about them as very large segments. But this is all about segment-ology. You get your DNA segments from your ancestors – your Mother is an ancestor, and she gave you the ultimate segments – entire chromosomes. See the chromosome 5 example in Figure 1.
So where did your Mother get this large segment? She got it from her parents – your two grandparents – through a process called recombination. Read on…
Recombination and Crossovers
Here is a very brief overview of recombination and crossover for genealogists:
A parent takes parts (segments) of the two chromosomes from her parents, and creates one new chromosome which she passes on to a child. Basically, when recombination occurs, a parent starts with one of their parent’s chromosomes and then shifts, or crosses over, to the other parent’s chromosome. This recombination results in two segments separated by one crossover. This process may be repeated several times on one chromosome. We’ll talk more about the probability of recombination below.
Recombination is a very complex process that is the cornerstone of life and diversity. You can google “DNA recombination” for more, but this brief summary is all you really need to know for genetic genealogy. See the Figures below to see examples of how this works.
Three important points:
- After recombination, the new chromosome is exactly the same size as each of the two chromosomes which were used to form it.
- The segments are “heel-and-toe” – that is, they are adjacent. When one segment ends, the next segment starts. There is no gap between segments.
- The crossover point marks the point between segments from two different ancestors. You change from one ancestor to another at this point.
Grandparents
So let’s look at your maternal chromosome 5 at the grandparent level. That is chromosome 5 with segments from the two maternal grandparents. See the chromosome 5 example in Figure 2.
There is lots of information here:
- Segments from the two grandparents “fill up” the entire chromosome.
- The segments from the grandparents alternate.
- There are three segments and two crossovers (we will look more into the number of segments and crossovers below).
- There are no gaps between segments.
- These segments tend to be large, and the crossover points tend to be widely separated.
- Note: Only ancestors from grandparent 1 can contribute to the segments from grandparent 1. In other words the segments for grandparent 1 can only come from the ancestors of grandparent 1. Ditto for grandparent 2.
Crossover Points
OK – before we continue, we need to look at the realistic number of segments and crossover points we should expect in each generation. Science has found that in one generation (Mother to you, for example), there are about 35 crossover points spread out over all 22 chromosomes. In fact the cM is defined by the probability of a crossover – such that there is a probability of a crossover every 100cM. So let’s look at Table 1:
Each atDNA testing company shows a slightly different table of cMs for each chromosome. You can see a report of cM for various companies at http://www.isogg.org/wiki/CentiMorgan. Don’t get hung up on the exact numbers – it’s the overall concept that counts. The average number of crossovers is the cMs in that chromosome divided by 100. Since the number of crossovers per chromosome must be a whole number, I’ve shown several alternatives above. Although the average may be 1 or 2 or 3, possibilities may include 0 or 4 (or more sometimes). The point is that there are only a few crossovers expected for each chromosome, in one generation, and if more occur on some chromosomes, there tends to be fewer on some other chromosome. This process occurs in each generation – for instance when a parent recombines the grandparent’s chromosomes and passes a single chromosome to you. It also happens when a grandparent recombines the Great grandparent’s chromosomes and passes a single chromosome to your parent. So there are about 35 crossover points at each and every generation, and they add up, depending on which generation is under consideration. This will be described for each generation in more detail below.
Important points from the above info:
- Recombination does not “puree” the DNA into tiny pieces (segments). Recombination tends to divide each chromosome into a few segments.
- Clearly with only a few recombinations, the resulting segments in one generation will tend to be large.
- Clearly with only a few crossover points, they tend to be distributed over the chromosome.
- DNA is random, and the number of crossovers in your chromosomes may vary. If you have more than average on some chromosomes, you will probably have fewer than average on some other chromosomes.
- Regardless of the number of crossovers, or their locations, all the resulting segments on a chromosome will fill the chromosome, with adjacent segments, from one end to the other.
- When there is 0 crossover, this means there was no recombination. This means that chromosome was passed intact to the next generation. Given the probabilities in Table 1, there is a high probability that at least one of the smaller chromosomes will be passed intact with each generation.
Great Grandparents
So, given the probability that there are two additional crossovers in each generation for chromosome 5, let’s look at a probable scenario from the Great grandparent’s perspective in Figure 3.
Important information from Figure 3:
- Again, segments from the four Great grandparents “fill up” the entire chromosome.
- The “new” segments from the Great grandparents alternate; and they alternate within their respective child. That is Ggp1 and Ggp2 are parents of grandparent 1; Ggp3 and Ggp4 are parents of grandparent 2.
- There are two new crossover points (shown by large vertical lines); and the previous crossover points are still there.
- There are no gaps between segments.
- Again, these segments tend to be large, and the crossover points tend to be widely separated.
- Note: Only grandparent 1 ancestors (Ggp1 and Ggp2) can contribute within the segments from grandparent 1. In other words the segments for grandparent 1 can only come from the ancestors of grandparent 1. Ditto for grandparent 2.
- We only had two new crossover points for chromosome 5, so they could only subdivide two of the three grandparent segments. Sometimes there may be 3 crossover points, but then, sometimes there may only be one crossover point. Even with 2 crossover points, they could have both occurred within one grandparent segment. When dealing with random DNA, there are many possibilities. We used the average of 2 crossover points to paint the best overall picture. You are invited to print Figure 3 and randomly place one, two, three or four crossover points anywhere you want. If you put two, or more, crossovers in one grandparent segment, be sure to alternate the Great grandparent’s segments: Ggp1-Ggp2-Ggp1…
- Note that there is no crossover point through the last segment for grandparent 1. That means the second grandparent1 segment was passed down, intact, from one of the Great grandparents. There is a 50% probability for either one. But only for one. We sometimes refer to such a segment as a “sticky segment”, because it appears to stick together through a generation.
- We now show five, rather large, segments at this Great grandparent level of chromosome 5.
- Note that the first new crossover point divides the grandparent 1 segment into two segments, one for each parent of grandparent 1. On the other hand, the second crossover point is separating two segments from different parents of the grandparents. These two parents, labeled Ggp2 and Ggp3 are not related to each other by marriage or otherwise. So some adjacent segments may be for husband and wife; and some may be for distant, unrelated ancestors.
2Great Grandparents
OK – moving on to the next generation back – continuing our Bottom-Up look… Again, we will add two more crossover points to get Figure 4:
Important information from Figure 4:
- As always, segments from the 2G grandparents “fill up” the entire chromosome.
- The “new” segments from the 2G grandparents alternate; and they alternate within their respective child. For instance 2Ggp5 and 2Ggp6 are parents of Ggp3.
- There are two new crossover points (shown by large vertical lines); and all the previous crossover points are still there.
- There are no gaps between segments.
- Again, these segments tend to be large, and the crossover points tend to be widely separated. But in this example the first new crossover point occurs fairly close to an existing crossover point, so a relatively small segment is created for 2Ggp3. It happens… and small segments are created.
- Again, only segments from parents can contribute to a child’s segment. So 2Ggp1 is a parent of Ggp1; 2Ggp3 & 4; are parents of Ggp2; 2Ggp5 & 6 are parents of Ggp3; 2Ggp8 is a parent of Ggp4; and 2Ggp3 is a parent of Ggp2. Note that there are no segments for 2Ggp2 or 2Ggp7 shown. Those ancestors did not contribute any DNA to chromosome 5.
- Note that there are no crossover points through the first, fourth and fifth segments at the Great grandparent look. That means these Great grandparent segments were passed down, intact, from one of the 2G grandparents. There is a 50% probability for either one, and I selected one of those in each case for example. Now we have two “sticky segments” from the previous generation; and one “sticky segment”, 2Ggp3, which survived three generations.
- We now show seven, mostly rather large, segments at this 2G grandparent level of chromosome 5.
- Again, note that the first new crossover point divides a segment into the two parents.
3Great grandparents
Let’s look at one more generation – to get the hang of it – and then draw some general conclusions about what to expect.
Important information from Figure 5:
- As always, segments from the 3G grandparents “fill up” the entire chromosome.
- The “new” segments from the 3G grandparents alternate; and they alternate within their respective child. For instance 3Ggp1 and 3Ggp2 are parents of 2Ggp1.
- There are two new crossover points (shown by large vertical lines); and the previous crossover points are still there.
- There are no gaps between segments.
- Although at this generation going back (Bottom-up), these segments tend to be large, but with each generation, a few segments are split into smaller segments. Another relatively small segment has been created.
- Now there are 8of the 16 3G grandparents missing (3Ggp3, 4, 5, 8, 9, 13, 14, and 15)
- The last, 3Ggp6, segment has now survived, intact, from the 3G grandparent level down to you.
- We now show nine segments at this 3G grandparent level of chromosome 5.
- Again, note that the first new crossover point divides a segment into the two parents.
So what are the big-picture observations:
- At each generation going back, each chromosome is made up 100% by segments from that generation.
- The segments at each generation are adjacent to each other; there are no gaps.
- On, average, there are only two new crossovers at each generation. So only two segments are subdivided at each generation.
- From here on out, at each generation, most of the segments will remain the same size; and only a few will be subdivided.
- Some segments, particularly the smaller ones, will appear to be “sticky” and survive for several generations without being subdivided.
- More and more ancestors, at each generation, will drop out of the picture as you move to more distant ancestors. This applies only to the chromosome under consideration. These ancestors may well be found on other chromosomes. Because the DNA is random, many of your ancestors will be represented on some chromosomes, for many more generations.
- This should dispel the common idea that all segments are cut in half with each generation. This may be true for averages, but in practice, we found above that only a very few segments are subdivided each generation.
- “Sticky segments” are normal. In fact, in dealing with the smaller segments and comparing with a parent, you’ll often find you have virtually the same segment as your parent, or none at all. More on this in a later blog post on shared segments.
- When a husband and wife have adjacent segments, then that crossover was created in their child.
- Although this “picture” was developed for Mother, the same principles apply to the father’s side of autosomal DNA. And, of course, everyone’s version would be uniquely different. But, on a big picture level, it would be somewhat similar.
- You can use Kitty’s chromosome mapping program to show your results at any generation up to 20 ancestors. Just list the ancestors of that generation in the MRCA column. See http://kittymunson.com/dna/ChromosomeMapper.php
Final Thoughts
Remember this whole discussion is based on your ancestral segments. Your ancestral segments are defined by the crossover points. The crossover points are locked into your DNA when you were conceived. They never change. They define the picture of your segments in each of your chromosomes. They define which ancestral lines contribute to which segments on your chromosomes. We’ll talk about this more in discussions about shared segments with Matches. We don’t really see the picture of our own segments in a chromosome browser. The browser doesn’t know where your crossover points are. What we see in a chromosome browser are shared segments. By grouping and Triangulating these shared segments, we can learn where the crossover points are. Much more in future blog posts….
05A Segmentology: Segments: Bottom-Up by Jim Bartlett 20150523
Epilogue
In my spreadsheets and analysis, I use Ahnentafel numbers. They are a standard numerical code for each ancestor: I am 1; my father is 2, my mother is 3; my 4 grandparents are 4-7; etc. They offer a unique shorthand for indentifying ancestors. Here is a summary of the chromosome 5 charts in this post, using Ahnentafel numbers for ancestors. Starting with 3 for my mother…

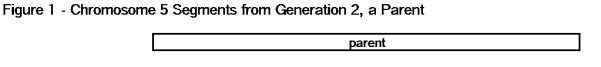
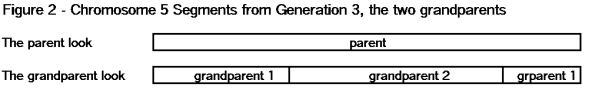





Jim, I want to check my understanding of what a DNA segment is and how many should be discoverable using triangulation. Referencing Figure 6, for example, we have one segment in the “parent look,” three segments in the “grandparent look” and nine segments in the “3G grandparent look.”
Each segment in Figure 6 has an ahnentafel number that identifies the ancestor who contributed DNA to downstream generations. My understanding is that each segment at each generational level could form the basis of a triangulation group. Is this correct?
In Figure 6, the number of segments is an arithmetic progression: 1, 3, 5, 7, 9 . . . because each generation adds two more crossovers. In principle, the number of segments could grow without bound into the thousands, millions, and even more!
On the other hand, you have written that 300-400 is the typical number of segments that one might expect to discover using triangulation. Why such a finite number?
The genome is 3400cM. If the threshold for detection of a triangulated group is 7cM, then is 3400cM/7cM = 485 is a reasonable estimate for the number of identifiable segments. If the threshold is 15cM, the number drops to about 227. Do these ballpark calculations perhaps explain why 300-400 is the “typical” number of segments that can reasonably be discovered using triangulation?
I am sorry for my belated reply, but your early posts are very helpful to check one’s understanding! Thank you again.
LikeLike
Andy, The 1-3-5…. applies to Chr 05. Chr 01 will have more crossovers and Chr 22 will generally have only one, sometimes 0 crossovers. Generall there is one chrossover for every 100cM… Your 227 vs 485 simple math analysis is spot on. The other thing to consider is the smaller the segment, the more distant the Common Ancestor – so we actually don’t want really small TG segments – the CA would be out of a genealogy time frame. Another way to look at it is that we use 7cM as a bare minimum segment size (any lower really increases the risk of false segments). And in general, due to random DNA, all the segments in a TG won’t be the same size (say 7cM) and stacked up like a pile up. They tend to be larger and more spread out – so using 7cM as a lower theshold, we tend to get the full TG somewhat larger. In fact, in practice they form in a wide range. If you start off with a 20cM threshold, you’ll get fewer, larger TGs. I think using around 10cM will be a good thereshod – but on the other hand everyone’s “mileage” will vary… Jim
LikeLike
Andy, I should have added that at each generation only 34 new crossovers are added. So the total number of segments from a parent is 23 – and they are big. The total from the grandparents is 23+34 or 57 segments (divided by 2 grandparents and spread over 23 chromosomes – so still pretty large segments (on average). We get 57+34 or 91 from our 4 Great grandparents on one side) – and (on average) they are smaller… etc. Jim
LikeLike
Thanks, Jim. I think I got you right when you said that after 3G grandparents, most segments will not get divided. Is there a cM value to go with that?
And, how widely, in your experience, do crossover points vary amongst siblings? I have several cousins plus uncle to play with (so most of 3 siblings), but a lot of the crossovers seem to vary very little–am I looking a data processing phenomenon?
Thx!!!
Kate
LikeLike
Kate – We all need to be very careful about establishing “rules” or even “rules of thumb”. We could easily do the simple math and say grandparents passed down 1/4 of their DNA to us – and work back up the generations, dividing by two. But Blaine’s Shared cM project shows us there is a wide distribution of values (spread over a curve) for each cousinship. I see a lot of segments in the 7 to 15cM range that are *usually* the same between my father and me. And some in that range, my father has and I got 0 (I got his DNA from his other parent). When I get up to 30cM and more I *tend* to see more partial segments (rarely exactly half) compared with my father. The key is to think an average of 34 crossovers in a genome (side) each generation – they are more likely to fall on the largest segments from the previous generation, but that doesn’t always happen that way. My advice, work with the data given to you, and see what it tells you – do not try to predict, but generally note if the result is reasonable. Jim
LikeLike
Kate, re: crossover vary amongst siblings… I’m not sure how to answer that. On the one hand each of the siblings gets their DNA from the same parents. Each parent already has their DNA etched in stone (so to speak) – they have many crossover points already fixed. When they pass their DNA to a child, they add an average of 34 more crossovers to the mix. These 34 new crossovers will be randomly different for each child. So it’s a mix. Each child will get 34+22=56 fairly large segments from their grandparents – these 56 segments will be random, while still covering the entire genome. However, the more distant crossovers will be the same for children who get the same grandparent DNA. In this respect, the great grandparent crossovers will be the same within, if the child got that part of the DNA – remember each child gets roughly 1/8 of their DNA from each great grandparent – some of it will overlap with other siblings, some will not. In summary, among siblings, it would be rare to get the same randomly generated crossovers created by the parent; siblings *will* get some of the same crossovers created by the grandparents; and I’d expect the number of crossovers from more distant ancestors to decrease with each additional generation. When you build a Chromosome Map based only on grandparent segments for each sibling, you can then see the same segment overlaps between any two siblings and can be confident that all the crossovers from prior generations (from the same side) will be the same within those overlaps. Hope this helps, Jim
LikeLike
Wonderful explanation of how each generation of cross-overs slices a chromosome into smaller & smaller segments. It illustrates how there is a certain orderliness about how cross-overs work. Cross-overs occur only within prescribed limits, and the resulting parts are NOT shuffled.
LikeLike
Andy, Thanks for your kind feedback. Your observations are good ones. Although cross-overs are random, they are not willy-nilly. Another key take-away is that every segment is not cut in half (a popular view that is not correct). Also, as the segments from an Ancestor get smaller, they are more frequently passed to the next generation intact – this leads to what looks like “sticky” segments that may persist for several generation. All things to look for and not be surprised about in our segments. Jim #IGHR2021
LikeLike
I am trying to figure out if I am genetically “more’ my father. or my mother? Is there a way to tell which is more genetically “me”?.
LikeLike
You got 23 chromosomes from your mother, and 23 from your father. There are about 22,000 genes – each parent gave you the same 22,000. From a DNA point of view you got almost the same from each parent.
LikeLike
Genetically, you are equal parts from you mother and your father. It’s the same “structure” from each, the same number of chromosomes and genes. But the genes from your two parents may well be different. Say one parent gives you brown eye DNA and the other gives you blue eye DNA. Which eye color you wind up with is a complex process, but you only wind up with one color, even though you carry both colors in your DNA. Your body only “expresses” one of the colors. To find out which of your 22,000 genes is “expressed” (i.e. used by your body), you’d need to catalog all 22,000 of yours, and your parents. Many of them would be the same – so it doesn’t matter, and neither parent gets the “credit”. But if they are different, then you’d need to figure out what they do and how to tell which one you are using. And rarely is there a one-to-one link between a single gene and some physical trait you can observe. So, I’d go back to the original statement: you are equal parts mom and dad.
LikeLike
Thank you for your responseJim…I have just discovered that I have a living, biological father. The DNA results came back with different cM…and I just wondered if I was genetically more HIM..or my mother because his numbers are slightly different. Your explanation helps..thank you. I guess I am just curious if I carry ‘him” in who I am..and I guess i do. Thank you . The funniest thing about emailing with you is that HIS name is Bartlett …(smile).
LikeLike
Deb – I sent you an email about BARTLETTs.
LikeLike
Fanstastic explanation of this unreachable mystery. Love your easy explanation. Thank You.
LikeLike
Thanks, Ralf – my goal is easy to understand explanations – so I appreciate your feedback. Jim
LikeLike
Hi Jim.
I am somehow related to a family of Swaffords. The problem is, their community was small and I have many cousins marrying cousins and related families. It is my understanding that the total shared CM in these lines makes my relationship look closer than it actually is.
I am wondering is there a way to estimate how closely I am related based on the overall length of the longest segment?
For instance, I share 75CM with two related people. One I share a 19CM segment and the other a 43CM segment. I assume that the person I share the 43CM segment with is more closely related to me, and the other might be someone I share with that is matching on mulitple lines.
Any ideas?
LikeLike
Lance, Our DNA is pretty random – so it’s not like solving an algebra problem, or surveying land etc. Roughly 1 in 10 true 3rd cousins will not show up as a Match at all. However, in general, what you assume (a larger segment is usually a closer relative, than a smaller one) is probably the way it is. One idea is to use the ISOGG/Wiki Shared Segment chart for the largest segment you share. In general, endogamy does not change an individual segment. Instead, endogamy results in additional shared segments (usually) which is why the total gives a skewed look. Another idea is to use Triangulation – each shared segment came to you down one specific ancestral line. The people who match you (through a shared segment), will have a Common Ancestor with you, somewhere on the same ancestral line. In other words, some Ancestor had to pass down the DNA segment to you and to your Match – you’re trying to find that Ancestor. With Triangulation, all of your Matches in the Triangulated Group who got roughly the same DNA segment, got it from the same Ancestor – down a specific line of descent to you (and a different line of descent to each Match). You may have that Ancestor in your ancestry is several different “boxes”, but only one of those “boxes” is the one the DNA came from – AND you will only have certain close cousins on that line, that you don’t have on the other instances of that Ancestor in different “boxes”. Hope this helps. Jim
LikeLike
Jim, I just have to say… I’ve been looking at your blog on and off for a few weeks, gradually wrapping my head around segmentology. You’ve done a wonderful thing with this blog. THANK YOU for the work you’ve put in, and for sharing it with all of us.
But THIS post… this post answered SO MANY of my questions, in one fell swoop. It makes so much sense now. I am energized and ready to make sense of it all in my own genome. Thanks again.
LikeLike
I am trying to wrap my brain around this subject and have a quick question? in the grandparent’s look in Figure 6, you show 7, 6, 7. Why is it not 6, 7, 6?
LikeLike
Gregory, It’s arbitrary. This is just an example – you are free to sketch your own example, the point is that at the grandparent level, for this chromosome, there will be about two crossover points – you can place them anywhere you want – they are random. Then if you have two crossovers, you have three segments – which will alternate between the two grandparents. To sketch an example you are free to use 7-6-7 or 6-7-6 on this maternal chromosome. On the paternal chromosome, with two crossovers, you could show 4-5-4 or 5-4-5. Just remember in the preceding generation – for Great grandparents – to use the correct Ahnentafel numbers. In other word if a 7 segment is divided in the next generation, it will have to be 14-15 or 15-14 – your choice for an example. In actual chromosome mapping there is only one correct solution. And it will not be the same as the hypothetical example I show – it will be different and unique for each person. Jim
LikeLike
Steve, In general, if the start or end of shared segments with close cousins line up, it will be a crossover point – but that is not a hard rule. As you Triangulate more and more Match/segments, the crossover points come into sharper focus.
LikeLike
One final quick question Jim and then I will get on with the Segmentology work and leave you in peace…………. I think I understand the mechanics involved around the snp , cM and mismatch bundle limit controls. If we lower the water level for a match you significantly increase the probability of an IBC event. However, is this still true for first cousin matches, which are already confirmed IBD. Are there issues in the measuring process of the DNA which will be carried forward at lower levels, even with first cousins? What I am thinking is whether I could ‘safely’ drop the limits to some level 300snps, 3cM (FTDNA record 2cM ‘matches’ ) between a one on one comparison between two first cousins and create a ‘better’ framework of close in family segments. Then add the triangulated segments from other ‘unknown’ matches at a reasonable match criteria 700snps, or 7cM (or higher, I believe a 12cM match criteria would be around 95% confidence in IBD). Am I adding noise or extra detail between first cousins is the question? I was thinking of going higher on a match criteria initially in the ‘unknown’ triangulation kits and see what the initial framework looked like for crossovers and then introduce more triangulated kits at a lower match criteria over time. Always moving from a position of near certainty (as much as we can), or is this a bit over the top !!
LikeLike
Steve, Working from large segments with closer cousins is a great way to start – get an initial framework established. Then drop the threshold and add more. Be prepared to subdivide some of the larger TGs.
I’ve tried lowering the threshold on close cousins, and it’s hit or miss – it appears to be random – so not a good plan IMO. When you get below 5 or 6cM the GEDmatch process generates too many IBC (false) segments. A big waste of time IMO, and it is certainly not a good basis for setting up other TGs.
Jim
LikeLike
Jim, I am having a problem with how first cousins overlap each other on average by only 880cM. If I do a Tops down from grandparents each grandparent passes 3,600cM in one chromosome to their child (lets call him parent 1) who is the parent of a grandchild (grandchild 1). Each of their children (Parent1 and Parent 2) have 50% overlap so have common DNA, on average about 1800cM from each grandparent. On GEDMATCH this would be recorded on average as 2,600cM on average rather than 3,600cM for both grandparents due to the fact that it does not account for the places where the two siblings have double identical segments across both chromosomes in a pair. The children of the siblings would have their parents DNA diluted by 50% from their parent’s spouses DNA. That means that the common DNA between the two parent siblings will be reduced by 50% so move from 1800cm to 900cm from each grandparent. So on average their should be 1800cM collective DNA from both grandparents shared between two first cousins. If you account for double identical positions this may look like 1300cM. So why do we only see 880cM as an average for Cousins as a complete overlap of both grandparent’s DNA. Can you see where I am going wrong ?
LikeLike
Steve, You started out right, but left out one step. Say 3600cM from grandparent to each child with 50% overlap between children is 1800cM. Or for both parents 3600cM (looks like 2600cM in browser). Now passing down one more generation is halved again to 900cM or a total of 1800cM from both grandparents – we agree up until now – but again there is only 50% overlap, so only a net of 900cM is shared between cousins. The nominal share between 1C is 1/8.
LikeLike
Thanks Jim I can see the error now, it has been driving me crazy !!! This is a great information resource, I have learnt so much from it. The biggest revelation for me was the small number of crossovers on average per chromosome. I am wrestling with that and the information you provided to work through the overlap logic to highlight the crossovers between 3 first cousins and one niece, daughter of a fourth first cousin all from the paternal line. In particular if 3 or 4 cousins line up at a specific position I assume it follows that since they all have that transition it must be an established crossover point from a common descendent, at least their grandparent but could be earlier. Thanks again for helping out above.
LikeLike
Hi Jim,
Please would you help me with some basic concepts – a bit of clarification as I think I have understood this blog.
Should one should see two sets of crossover maps for “me”, which comprise 22 from paternal and 22 from maternal? I assume my two “strands” for each chromosome pair are one entirely from my mother and one entirely from my father and there is no crossover or mixing up between the two strands whatsoever. Following on and if this is right, the cross overs occur only within one chromosome strand from, for example, the maternal grandparents only, if it is the maternal strand from the pair under consideration and vice versa for the paternal strand. Is that somewhere near the mark?
I’m looking forward to “There will be more about shared segments in later other blog posts.” It seems to me that my matches are from, for example, the maternal between location a and b on a chromosome and then straight after a block from the other side. Occasionally there is a “B” side of overlapping segments, but this is rare, Perhaps I simply do not have enough matches in my spreadsheet yet.
All the best and thank you very much for your informative blogs – they are a great help!
David
LikeLike
David,
Yes, your maternal and paternal crossover maps are independent. Each set of your chromosomes was independently created in each parent before you were conceived – there could be no mixing between your parents. And, yes, on one maternal chromosome, segments from your two maternal grandparents are adjacent and alternating. NB: An entire chromosome could be from only one grandparent – one large segment. In fact there is a high probability that this happened in at least one of your shorter chromosomes (17 to 22).
LikeLike
Another researcher told me I could get lucky and have it all triangulate on the maternal Armstrong strand. I find it hard to get my head around with what appears one huge segment which at this point seems to overlay all the Brady segments, especially with the mix of surnames in the paternal and maternal lines. That is why I have been re-reading this post (several times). Am I missing something other than having my cousin upload to Gedmatch?
LikeLiked by 1 person
It helps to have as many shared segments, over 7cM, in the analysis. Your DNA has natural breakpoints at the crossovers – many segments help to define these points, IMO.
LikeLiked by 1 person
Barbara, The Matches are pouring in – lots of data to work with. And, yes, some of them are in conflict – it’s because I have so much Colonial Virginia ancestry and it’s easy to relate to Matches several ways, and on both sides. But at the end of the puzzle, all the pieces should fit – there is only one correct solution!
LikeLiked by 2 people
It seems to me that descent with endogamy is more like a braid than a tree. I would like to create a 3D representation of this, maybe a sort of macrame sculpture! It’s possible now with computer charting to create beautiful 3D graphs, although I don’t have the skills yet – I am struggling with learning how to create a stacked bar chart!
LikeLike
Mizmdk, the easiest way—(I’m all about easy) donate $10 to Tier 1 go to Triangulation Groups Beta. Enter you kit number, put on a pot for tea. Spend a few minutes sprucing up the kitchen, or hanging up clothes (assuming you are usually too busy doing genealogy to spend much time on other stuff). When the results appear, click on Segment Bar Chart, copy, paste into a spreadsheet. Do the same for the Triangulated Graphic Tree and the same for the csv file. Enjoy a cup of tea while you look over the Segment Bar Chart. See all those green bars stacked up neatly? They indicate common ancestors. 🙂
LikeLike
Okay, I have this. I do not have my maternal 1/2 first cousin’s full tree to view yet. To restate our shared line is through our maternal grandmother. My half first cousin Armstrong recently had DNA done. In the meantime, I have been identifying my segments as I can. I have been concentrating heavily on a specific family in my paternal Brady line. I was curious about some surnames from my grandmother’s line that were showing up (Green and Justice). I decided it would be a good idea to enter all my half first cousin’s DNA so I could better define my grandmother’s line. Surprise! I ran into a surname from my maternal grandfather’s line (Hohimer) to add to the mix. On Chr 12 I was excited because it is one chromosome where I have made loads of connections on the Brady family in my paternal 5th great grandfather on my father’s maternal line. I thought entering my Armstrong cousin’s areas where we match would help me define recombination areas. My half cousin, not yet on Gedmatch, I transferred the results from FTDNA. We share a whopping 95 cM on chromosome 12 and it is smack dab in the middle of the family matches in my paternal Brady line. I have found two surnames (Justice and Greene) I have walked back in my maternal grandmother’s line which I suspect are part of the reason we share such a large segment on this chromosome and have done my best to expand my tree to answer to address this. I believe your suggestion of TG’s is the answer. With the information on his grandfather’s line I actually believe I can do this; even though some of the segments in the TG’s might be quite small. What a wonderful puzzle. It like a beautiful assortment of silver and gold chains tangled in the bottom of a jewelry box. Patience, persistence with TG’s acting as the oil to help them easily slide apart. Thanks for the advice! Endogamy though a bit of a headache at times, can be fun! Still having a blast.
LikeLiked by 1 person
Pingback: Using a Child to Determine the Side | segment-ology
Jim, I am re-reading this and my dimmer switch turned low (again). Where is my thinking mis-firing? I am working on a family project. We are all doing TG and segment studies. We are focusing on a common set of great grandparents. For me they are my 5th great-grandparents. I am coming up with loads of matches who also match this couple. How do I determine which chromosome is from my fifth great grandfather and which is from my 5th great-grandmother when we all share both in our trees? I am sticking with chrs over 15 cMs. This couple had 9 children. I am matching with descendants from four of those nine children so far. I have tried comparing with the maternal surname and though I am finding matches, I have not yet been able to define who her father was as she is not showing up in any of the paper trails of those with the same surname who lived in the same area (we’re talking walking distance) at the right time.
LikeLike
Barbara, When focused on 5th Great (5G) grandparents – this is the 6th cousin (6C) level, and you might have more than one Common Ancestor with each of your Matches. So, the best approach, IMO, is to try to find intermediate cousins, to be sure to “walk the ancestor back”. I am working on a similar study of my HIGGINBOTHAM ancestors, at the 5G to 7G grandparent level (6C to 8C). I have found close to 100 of my DNA Matches who have this line in their ancestry. I now have about half of them at GEDmatch and/or FTDNA. It turns out that one group of them is in one Triangulated Group (TG) (they have matching, overlapping, shared segments). In two other cases there are 2-4 Matches in each TG, making three different TGs (on different chromosomes) with multiple HIGGINBOTHAM Matches. The rest of my HIGGINBOTHAM cousins, are lonely singletons, each in a different TG. Although they may be genealogy cousins, I don’t expect the DNA segments in their TGs to come from a HIGGINBOTHAM ancestor – the DNA probably comes down a diffent line. Actually, this is expected. At the 6G grandparent level (7Cs), we have 128 couples on each side (HIGGINBOTHAM is on my maternal side). Based on my experience mapping TGs over 95% of my DNA, I know there are about 200 TGs on my maternal side. So I’d expect, on average, about 2 of them for each of my 128 couples – some will have 0 TGs, some 1 and some 3, or even 4, but most ancestral couples at that distance will have about 2 TGs. Remember: the distribution of DNA from ancestors, and the number of TGs for each, depends on the randomness of DNA, and not on family size, ethnicity, geography, etc. factors. Also remember: you cannot tell which partner in a TG CA passed down the DNA, unless you can find/confirm Matches in the TG who go back another generation.
So bottom line for your quest: I would group all of your appropriate cousin/Matches by TGs, and focus on the ones which have the most Matches.
Hope this helps, Jim
LikeLike
Hi Jim, thanks for this great blog. Things are beginning to clear up for me. There is one bit of confusion for me, I don’t understand the 59 segment on the right end of the 3G-GP chromosome. Should that be a 56 or 57?
LikeLike
Jim, You are correct – my error – good catch!
LikeLike
Pingback: First 1st Cousin DNA Results: Part 2 – Trying to Explain Aunt/Nephew Matches – Hartley DNA & Genealogy
Pingback: Understanding and Using TGs | segment-ology
My brother tested Y-DNA in 2008 37 marker he matches 4 Rock surnames, and 1 Bartlett John Joseph P Bartlett/Barrtlett b 1909 (was adopted) my grandfather was not adopted but raised by foster parents b abt 1868 Indiana no records; I have done the autosomal DNA fmtree-June- 2016 what I am asking here this, will I also show a Rock and Barnett connection? I have found none so far but I have 6 matches on segments 4 @ 19 1 @ 20 and 1@21 just what does this mean?
Also I have a connection with Marilyn Hutton her gr grandfather and mine are brothers- we have good paper (proof) but yet we have not connected with DNA; why is that?
LikeLike
Eleanor, I would take it as a strong clue that your paternal surname is ROCK. About 1/2 of your autosomal DNA Match are probably on his side and half on your maternal side. You will match others with a blood (biological) relationship – that’s the path the DNA follows. You should get not real matches with any foster parents (unless there is some distant blood relationship). When starting out with autosomal DNA, I would work with (contact and shareinfo with) your closest Matches first – they are the ones who are listed first.
If you don’t share any DNA segments with a known 2nd cousin, it may be that there is a break in the blood line – either on your side and/or on Marilyn’s side. I would highly recommend you both register at http://www.GEDmatch.com and upload your raw DNA files there. You can then get an independent check of a Match – a 2nd cousin should almost always match.
LikeLike
Thank thank you, another thing do you think I should get a mtDna test would that help to make further away connections on the female lines?
Ellie
LikeLike
Mtdna is not generally successful when fishing for relatives – it seldom works out. You’d probably get some matches, but they’d probably be 10-30 generations back. My recommendation would be to get atDNA test for your closest cousins – they’ll help sort your Matches out.
LikeLike
One question, my daughter has 3-4 chromosomes where the entire segment matches only one of my parents. I understand the crossover, but I guess I don’t understand why one parent would give all the DNA, though my mother, cousin and I all share an entire chromosome in common as well.
LikeLike
Jennifer, A parent has two of each chromosome – they “recombine” these into two new chromosomes and one is passed to the child. The recombination process usually involves one to three crossovers (resulting in two to four large segments), but sometimes there are no crossovers (resulting in one of the two chromosomes being passed down intact), or four or five crossovers (resulting in more, but smaller segments). On average, we should expect one or two of the chromosomes to be passed along intact in each generation. Usually it will be different for each child, but it doesn’t have to be.
LikeLike
Pingback: My Father In Law’s Grandparents’ DNA – Hartley DNA & Genealogy
First of all I’d like to tell you that I’ve loved all of your posts! They’ve helped me tremendously and I re-read them quite often. I was reviewing Segments: Bottoms-Up and have been trying to break down my chromosomes into these segments. I’m hoping you might clarify something for me as I want to ensure that I’m not making errors from the start. Just when I think I understand this, I get a bit jumbled up.
My mother shares one 150 cM segment on chromosome 1 with her maternal aunt. Would it be correct to assume that this 150 cM segment came from only one of my great aunt’s parents?
LikeLike
No. An Aunt is too close – she shares a lot of DNA with your mother’s mother which came from their parents. Since they are so close, and have long stretches of identical DNA, it’s possible to get 150cM that is split between the parents of your mother’s mother and your mother’s aunt. In many respects we need to treat an aunt or uncle just as we would a parent. Perhaps think of parents with two children (you mother’s mother and her aunt) – they each get big blocks of DNA from their parents – some overlaps, some does not – but the part that overlaps, could be part from one parent and part from the other. Use triangulated groups to sort out the 150cM stretch of DNA.
LikeLike
Thank you so much for your reply and so quickly too! I truly appreciate it.
LikeLike
Pingback: Does Triangulation Work? | segment-ology
Kim
1. Your DNA came from 4 grandparents – roughly 1/4 of your DNA from each.
2. I’ve worked hard for 5 years to map my chromosomes to my 4 grandparents, and I still have 10% which is not mapped – I don’t know which grandparent will be linked to each of the remaining segments.
3. I doubt that your current Matches cover all of the segments from your grandmother.
4. The bottom line is you cannot assume the remaining Matches go to one grandparent.
LikeLike
Hi Jim…question for you..I have my grandmother tested..so can I safely say that any gaps in our shared chromosomes belong to my grandfather?..Thank you!!!
LikeLike
Pingback: The Porcupine Chart | segment-ology
Pingback: The Porcupine Chart | segment-ology
Pingback: Segments: Top-Down | segment-ology
Thanks for your clear description of cross-over points. I have one question please. Are the cross-over points on the chromosome you inherit from your father exactly the same as those on the same chromosome you inherit from your mother? For example, if the first cross-over point on my paternal chr1 is at location 13,100,409, will the first cross-over point on my maternal chr1 also be at location 13,100,409, and so on? Or is there no relationship whatsoever? Thanks.
LikeLike
Lennel – thanks for the question. The answer is that your maternal chromosomes and their crossover point are generated completely randomly in your mother, with no knowledge of or interaction with your father. The crossover points in each chromosome are independent and random.
LikeLike
I’m so delighted that you decided to blog. Thank you for sharing your knowledge of this complex topic! Like several others, I found this post regarding crossovers to be especially helpful.
LikeLike
Thanks for your encouragement.
LikeLike
Pingback: Fuzzy Data, Fuzzy Segments – No Worry | segment-ology
Im loving your blog very clear and easy to follow makes things so much better to understand. Thank you!!!
LikeLike
Kim, Thank you for the feedback
LikeLike
Thank you for your “Bottom Up” post. I have a much better idea of atDNA now, which should prove helpful as I compare my test results with my matches.
LikeLike
Diane,
Thank you. When you get some Common Ancestors worked out, try Kitty’s Chromosome Mapper to see the shared segments. For many that picture is a very helpful tool that highlight questionable areas which are probably more chopped up than you should expect.
LikeLike
June,
All close relatives are helpful. 1 – they usually share several segments; 2 – you already know the MRCA, which serves as a “pointer” for the CA of each TG; you also know the side for each TG they are in.
LikeLiked by 1 person
I only have one full sibling. She has been tested. I also have a half-sibling. How much would getting him tested help in determining crossover points given that he also has his mother’s DNA in the mix?
LikeLike
The diagram with Ahntafel numbers is a very clear way of presenting it/keeping track of it. Thanks!
And thanks for this comment in the reply on the “three siblings is enough” point…time to get that extra kit…..
LikeLike
Thanks very much for the clear explanation, which prompted me to closer at the bottom levels. I compared two full siblings (myself and my sister) to the phased paternal genome and noticed that the very large segments the siblings have in common break at about the same rate — 0 to 4 times per chromosome. Will these shared segments tend to start and end on the crossover points for the grandparents or are they more likely to be random?
LikeLike
With three siblings you can determine all of the grandparent crossover points. I had to do the diagrams to believe this myself. With only two siblings it may work for some.
LikeLike
Interesting! Now my brother is getting a kit for his birthday😀
LikeLike
That is interesting. I’d like to see the diagrams
LikeLike
Jason
This blog post includes one Table and several Figures. They are separate images inserted after the text. It appears OK when I publish the post, but many folks have different computers and software. Try using Google’s free Chrome browser.
LikeLike
I would love to see how this works. I have a sibling set of three as well as a number of first cousins on both sides. Would love to see how you might be ab,e to illustrate it. On my maternal grandmother’s side, her grandparents were all recent immigrants so finding tg groups is hard. I’m having a hard time conceptually pulling these siblingships apart into understanding where crossovers must be since they have areas of the chromosome where they match one parent, neither parent or both parents. I love this series and would love to see more with comparisons of siblings or an individual and close relation such as an uncle or first cousin.
Again, great work,
Mindy
LikeLike
Thanks, Mindy – stay tuned for more…
LikeLike
Wow! The light bulb went from completely out to dim. Thank you.
If I understand correctly, using chromosome 22 as an example. I may have 0 crossover points in which case I inherited the entire chromosome from either mother or father? With 1 crossover point, I may have father segment concatenated with mother segment OR mother segment concatenated with father segment? Can there be a case of chromosome 22 ever having 2 or more crossover points? I assume yes, but the probability is very slight.
Can’t wait to see how to determine the crossover points.
LikeLike
Yes, you probably did get one or more of chromosomes 19-22 intact from one grandparent. And yes it is possible to have two crossovers in one generation sometimes. NB: with each generation, you are likely to get additional crossover points on these small chromosomes, so when you are looking at shared segments with distant cousins, and Triangulating, you’ll determine your several/many crossover points on those chromosomes.
LikeLike
Thank you for that excellent presentation. It makes sense!
LikeLike
Thank you
LikeLike
Thank you, Jim. And, at some point will you address “pile-ups.”
LikeLike
Thank you. At some point I will address pile-ups, but the short answer is that pile-ups are not an issue with segments over 7cM which Triangulate. In fact, in this case, pile-ups are helpful – they give you many more cousins to help determine the Common Ancestor. Pile-ups with short shared segments may go either way – TG “jury” is still out.
LikeLiked by 1 person
Jim, thank you so much for sharing your in depth knowledge of this process in such a clear yet concise way. It has significantly advanced my understanding and will greatly advance my analysis.
LikeLike
Thank you
LikeLike
Thank you, this is a terrific summary.
LikeLiked by 1 person
Thank you
LikeLike
Jim, I can’t get the figures to load for me! I use Safari!
Thanks
Margo
LikeLike
Margo
Please try Chrome browser. I cannot seem to add graphics except as pictures.
LikeLike