In blog post Segments: Bottom-Up we looked at ancestral segments from the bottom up. That is we started with you and the very large segments (aka chromosomes) you got from each parent. Then we continued with two crossovers per generation to the 3G grandparent level. Of course, this is not the way DNA works! DNA comes from our ancestors to us – Top Down. So let’s see if we can reconstruct the segments we had in Figure 6 of Segments: Bottom Up. Here is that Figure again as Figure 1:
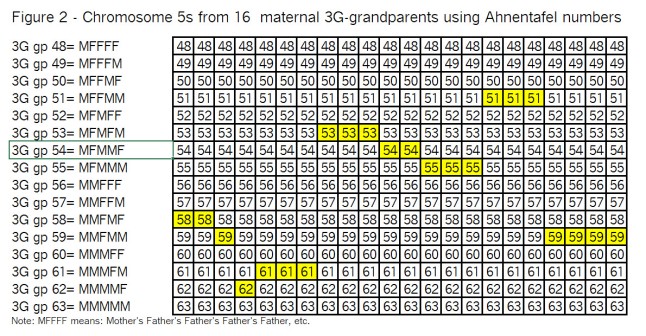
To do that, we have to start with the chromosomes of the 16 3G-grandparents; and take them Top-Down a pair at a time. Ready…? Here we go – Figure 2:
Observations from Figure 2:
I’ve highlighted the segments (in yellow) that we wound up with in Figure 1, just so you (and I ;>j) can keep track of them as we go through the recombination process.
Note that with only 2 crossovers per generation, there are not a lot of subdivided segments.
In the left-hand area, “3G gp” is Great, Great, Great grandparent; the numbers 48 to 63 are Ahnentafel numbers; and M means mother; F means father.
So the next step is recombination of each pair of parents, as show below. This mean 2 crossovers for each pair (noted by the vertical lines, and flipping the 2 chromosomes after each crossover. Then one of these 2 chromosomes is not used (noted by the X). See Figure 3:
Observations from Figure 3:
The highlighted segments are all in the chromosome which gets passed on to the child.
Note that at each generation, the highlighted areas “cover” the whole chromosome.
One of the two chromosomes is X’d as it is not passed to the child.
The remaining chromosome in each pair is passed to the child
The child is noted by an Ahnentafel number which is half – e.g. 28 is the child of 56 and 57.
Let’s see how this looks with the remaining ancestors in pairs in Figure 4:
As before these pairs of chromosomes are recombined at the crossover points as shown in Figure 5:
Observations from Figure 5:
Same as observations from Figure 3.
Compare these segments (Ahnentafel numbers and locations) with Figure 1.
Let’s see how this looks with the remaining 4 G-grandparents, arranged in pairs in Figure 6:
As before these pairs of chromosomes are recombined at the crossover points as shown in Figure 7:
Grouping these two remaining maternal grandparent chromosomes in the mother, for recombination seems very familiar from the Segments: Bottom Up blog post. Let’s see in Figure 8:
See if you can determine where the 2 recombination points have to be, in order to wind up with the same results we show in Figure 1 – which was the final result of a Bottom-Up look. Here is Figure 9:
Finally we have the chromosome from your mother in Figure 10:
Final Observations:
A comparison with Figure 1 shows the same outcome with a Bottom Up review.
There were many of the “random” recombination points above which could have been different because, in the end, that portion of the DNA was not passed down.
But certain recombination points had to be where I selected them (not at random), because I was working toward a known chromosome map – the one we came up with in the Bottom-Up review. If I had been truly random in the Top-Down analysis, I could go back and recreate the same answer using Bottom-Up. The point is that you can arrive at the same answer using a Bottom-Up or a Top-Down analysis.
Your DNA already has all of these recombination points in it. Triangulated Groups will help you define these points.
For you, each recombination point has already been determined, some at one generation and some at another. How to determine which generation it is will be a separate blog post… (when I figure it out ;>j – think MRCA with each Match…)
Remember there are about 70 recombination points created across all of your chromosomes in one generation – usually a few more from your mother’s side, compared to your father, but it’s an average, and DNA is random.
Note that your recombination points will not be the same as your Match’s recombination points – your Match almost certainly did not get the same ancestral segment you did (re-read What is a Segment? for the difference between ancestral segments and shared segments). This is one of the key reasons why spreadsheets (with shared segments) for different people should be kept separate. Although you may have a shared segment with a Match, there is no correlation between your ancestral segments/crossover points and your Match’s.
We can see above, the generation where a particular ancestor’s DNA is no longer included. We noted this in the Bottom-Up post as well. Although that ancestor is no longer on this chromosome, he/she could well be on a different chromosome.
Note that the ancestors whose DNA is no longer included on your chromosome, is not the result of segments being subdivided into oblivion (or even small segments that don’t show up as a match). Ancestors dropped out of the picture largely because their DNA was on the chromosome that was not used. At every generation, half of the DNA involved is not used! It’s no wonder, some ancestors drop out. Again, we are not talking about little pieces of DNA, but large chunks in many cases.
You are encouraged to take pencil and paper (or a spreadsheet), and try variations on crossover points and recombination. Play with crossover points at various locations. Put them close together in one generation to create a very small ancestral segment. Or try dividing segments in half with each generation (averaging, say, 2 crossovers per generation) and see how many generations there are before all the segments are below a threshold (say 7cM). I think you’d be surprised at how many generations back you can go. And if you unbalance the process (not cutting each segment in half (50/50), but say 25/75, or even 10/90), you will get even more generations with above threshold segments.
Have fun with it.
05B Segmentology – Segments: Top-Down by Jim Bartlett 20150601










This confuses me.
Above you say:
“Although you may have a shared segment with a Match, there is no correlation between your ancestral segments/crossover points and your Match’s.”
From How to triangulate.
“All of these segments are shared segments with you. So with segments that overlap each other, you want to know if they match each other at this LOCATION. If so this is Triangulation”
If there is no correlation between location, then how do you triangulate?
My understanding was that if I match someone on my maternal chromosome at a specific location and my cousin matches that same person at that location on her paternal chromosome, then we should both be seeing the same ancestor’s dna. It may have gotten chopped off at the beginning or ending because of crossovers but that shared segment will still have come from the same ancestor.
LikeLike
Susan – You are right, I am right and your understanding is correct. If you look at shared segments in a Triangulated Group, they almost never have the same start and stop locations. Your Match gets a segment from your Common Ancestor, and you get a segment from the CA – they are rarely exactly the same. And what you see in a Chromosome Browser is only the part that overlaps. And each TG will have a bunch of these shared segments that “fill up” a TG. The boundaries of the TG are crossover points, and all the DNA in the TG came from the CA – one large segment from the CA to you. Each of your Matches in your TG will probably have some of your Matches and maybe some other Matches and they will have a somewhat different TG location and large segment from the CA (they don’t get the same segment from the CA that you did). So the bottom line is the you have several hundred crossover points over all of your chromosomes – they are fixed. Each of your Matches will have different crossover points. Their TG segment from the CA and your TG segment from the CA will overlap – and the overlap will be within each of your TGs, but that one overlapping shared segment from each Match does not define your crossover points – only the ones at the two ends of a TG will. And remember: the ends of a TG are a little fuzzy. A better way I could word the Triangulation definition is to say: “they match each other within the TG boundaries.” The overlapping segments in a TG don’t all have the same start and end locations.
LikeLike
Hi Jim, thanks for you great detailed and informative explanations. I am confused. It seems that Ahnantafel 28 should have been 29 in your example.
LikeLike
Jim – again, my (consistent) mistake. Either change 59 to 57 or 58, or change 28 to 29 – I think you understand what I’m trying to show: crossover points are created at each generation; your DNA is full of them; but there are only and average of 34 per side per generation, so some segments are not subdivided in each generation, and are passed on intact (or “sticky”).
Jim
LikeLike
Pingback: Understanding and Using TGs | segment-ology
Jim, I have followed your blogs with much interest, and you have given me fresh new ways to do analysis on my DNA matches. Everyone recommends testing with different companies. I am willing to do that, if necessary. However, I have a question: I know that we do not know whether our results for a chromosome came from the pair from our maternal or paternal lines. Does testing with other companies offer the possibility of finding the other one of the pair? Or do they always test the same one? I seem to remember reading somewhere about testing on the “presenting” one. Could that possibly change from one test to another?
I was adopted, and know my birth families. I have no full siblings, and have found a half-niece on my maternal line who has tested. That line is going full guns on the DNA charts because of Mennonite ancestors. However, my paternal line is my brick wall. My 3-Ggrandfather is from Ireland, and even lowering the thresholds on Gedmatch, I don’t find anything (well, I do, but nothing matches up with the surname). I have paternal half-siblings (one male, three female) who can’t be located, or do not want any contact. I would have paid to test my half-brother on a Y-DNA test, but I can’t find him. I have proven DNA descent on almost all of the women who married into this line.
What do you suggest?
LikeLike
Cheryl, you get a pair of each of the autosomal chromosomes (1 to 22). You get one from each parent. And we normally try to figure out which one of the pair a Match is on – the maternal chromosome or paternal chromosome. We usually say the match is on the maternal side or paternal side – meaning which side of your ancestry, which is the same as which chromosome (maternal or paternal).
All three companies (FTDNA, 23andMe and AncestryDNA) use the same basic process. And that process cannot tell us which side (maternal or paternal) the Match is on. To do that you have to have some genealogy – which includes matching a close relative’s test. There is one deviation from this “rule” – if one parent has significantly different ancestry – say UK and Jewish, or Asian and African, etc. Then it sometimes works to look at the Matches (and maybe their ancestral surnames, and where they are from), to determine which side the match is on.
I also recommend testing at all three companies. Each one has a very large database of folks who have tested. At each company you are compared to only the folks in their database. There are many more Matches out there at the other companies. You never know where a close Match might have tested. You should also upload your results to http://www.GEDmatch.com – there you will get 2,000 Matches with names, emails, and DNA segment info for each Match. GEDmatch also has several different admixture utilities.
I suggest anyone, and adoptees in particular, should test at all 3, upload to GEDmatch, and do Triangulation (start with segments over 15cM or larger, then drop down to 10cM, etc.) Triangulation forms groups which make great research Teams. Also if you determine a group is on one side, overlapping groups will be on the other side.
Good luck,
Jim
LikeLike
Pingback: Endogamy PART II | segment-ology
Pingback: Does Triangulation Work? | segment-ology
Pingback: The Porcupine Chart | segment-ology
Pingback: The Porcupine Chart | segment-ology
Your blog posts are amazing! Thanks so much for sharing your knowledge with us.
LikeLike
Thanks for the kind words, Nancy – you made my day. Actually I learn more with every post I write – I have to think it out as carefully as I can, and then explain it the same way.
Thanks again,
Jim – Sent from my iPhone – FaceTime!
>
LikeLike
Hi Jim, I love reading your post and trying to understand, I understand how hard It must be to share your knowledge in a way the untrained person can understand. I must admit that I get lost in the scientific weeds. LOL I was wondering where the Ahnentafel numbers come from. Thank you for taking the time in doing this. Much appreciated.
LikeLike
Paul, Thanks for the kind (and encouraging) feedback. Please google “Ahnantafel numbers” for a fuller explanation. But in short: you are #1; your father is #2; your mother is #3; your 4 grandparents are 4-7; etc. The numbers are tagged to specific ancestors such that one ancestor’s father is twice their number and the mother is one number higher. Each of your ancestors has a specific number.
LikeLike