Shared IBD segments come from a Common Ancestor (CA). Matching & overlapping IBD segments form Triangulated Groups (TGs). Every Match in a TG with significantly overlapping shared segments will have the same CA! And closer Matches (cousins), will also have a closer CA. So how can we have a close CA and a distant CA when they are in the same TG? When they are all in the same ancestral line!
Let’s start with a distant cousin (Match) and look at the Common Ancestor.

Some notes about Figure 1
– With atDNA the path from the CA can go through males (boxes) and/or females (circles) in any order – it does not matter.
– The CA is one of the two parents above – the DNA that passed down from the CA to you and your 7th cousin (7C) came from one person. In this example, I’ve assumed the mother just to illustrate that it is just one parent. In most cases we don’t know which parent the DNA is from.
– The CA has at least two children: one is an ancestor of your 7C Match (M); and one is the ancestor of you (U)
– In this case the CA is also an MRCA (Most Recent Common Ancestor) – you and your Match don’t relate any closer on this line. However, in genetic genealogy, we tend to call this the CA, rather than the MRCA.
– You and your Match (M), will also share all of the Ancestors of the CA.
– This Figure 1 assumes the CA shown is the correct CA – the one who passed down the shared DNA segment to you and your Match. We don’t really know if this CA is correct, until we find corroborating evidence – read on.
So how do we confirm that this CA line (either the mother or the father) is the one who passed down the segment you and the Match (M) share (as opposed to some other ancestral line)? One way is by Triangulation. When several people share the same segment, and all have paper trails to the same CA, we assume this CA must be correct. Another method is by “walking the ancestry back”. That is through closer cousins who also share this segment (in a Triangulated Group). We are generally pretty comfortable (when a close cousin shares a lot of DNA with us) that the closer CA is correct. In other words, when a 2nd cousin (2C) shares 220cM with us, and has large individual shared segments with us, we assume the CA is the known Great grandparent. And with large segments this is almost always true. Then if a known 4C shares a good sized segment with us, we also assume the known 3G grandparent is the CA. If all of these occur in the same TG, we need to call the intermediate CAs, MRCAs (Most Recent Common Ancestors) to distinguish them from the CA of the TG. Let’s see how this looks in Figure 2.
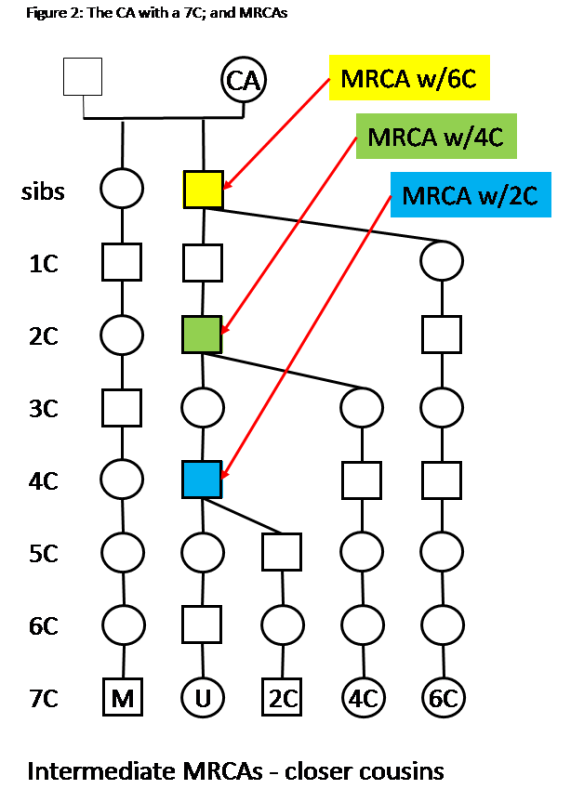
Some notes on Figure 2:
– The Tree for your Match (7C) and you is the same as in figure 1.
– A matching 2C on the same segment will have an MRCA with you on the G grandparent.
– Matches with 4C and 6C are also shown with MRCAs on the ancestral line from you to the CA.
– Everyone in Figure 2 descends from the CA.
This scenario, with intermediate MRCAs, adds a lot of confidence to the CA being the Ancestor who passed down the DNA that all of you (Match M, you, 2C, 4C and 6C) share.
Note that the intermediate MRCAs could have just as easily been on the Matches line. And/or you and the Match may both have intermediate MRCAs. The key point is that the MRCAs are in the ancestral line to the CA.
This concept applies equally to TGs. Each TG really represents a segment from an Ancestor to you. A “tight” TG – one with significantly overlapping segments among all the Matches in the TG – will have a CA just like a shared segment does. And all the Matches in the TG will share that CA. A “wide” TG – with “cascading” segments such that one at the beginning of the TG doesn’t overlap one at the end of the TG – may well turn out to be two TGs, with two CAs… more on that in a different post.
So there is always an Ancestor who is the most distant MRCA of your TG, for a given threshold. That means that any Ancestor who is more distant would not show up as a Match (using the given threshold, say 7cM), because the segment from the more distant Ancestor down to you would be too small at that distance to match anyone. For example, you may have gotten only 6cM from a 7G grandparent. In that case you would never get any Matches who were cousins on that 7G grandparent, using a 7cM match threshold. Others may get large enough segments from that same 7G grandparent, and maybe get some Matches, but you would not.
It appears this most distant MRCA of a TG may be fairly deep in our Trees in many cases. As a result we are having a hard time finding them. Our best tactic then is testing close cousins, and finding intermediate cousins among all of our Matches. This means testing at all companies and uploading to GEDmatch to get the most Matches you can. We never know when the key intermediate Match will show up – they won’t always have significantly larger segments. And lowering the threshold at GEDmatch, in general, will only result in even more distant cousins and more distant CAs.
Summary
A shared segment is from a Common Ancestor (CA) with a Match (cousin).
Closer cousins would have MRCAs with you who are descendants of the CA. Your Matches may also have closer cousins with MRCAs who also descend from the CA.
These “intermediate” MRCAs increase the probability that the CA passed down the shared segments.
We still do not know if the CA is the mother or the father, but we can be very confident that this is the correct ancestral line, and not some different or alternative ancestral line.
05C Segment-ology: CA and MRCA by Jim Bartlett 20160101

You mentioned vertical columns and cascading patterns formed by TM match groups, and I’ve been looking for a more detailed explanation, as to how matches can appear in either a vertical column or a more cascading pattern! I think (?) I may be seeing cascade formations where the MRCA for the beginning part of the TG group, is the ancestor of someone who married the descendent of the other MRCA which contributed segments further along in the cascade, and the cascade maybe represents different recombinations of these two grandparents DNA from different lines, which were passed on in different sized segments by the parents to the kids…? But my understanding of this is still really murky and I would love to know more about the differences in how vertical columns of matches and cascading patterns form. Is this explained in more detail in some of your posts? Doing a search for “cascading” isn’t turning up anything!
LikeLike
I mentioned “cascading” once in my blogpost. My point was that the segments in a TG are not all the same size and the ends are often at different points. I was trying to describe how this looks. See the diagrams in this post: https://segmentology.org/2016/02/05/anatomy-of-a-tg/
What we see in a TG are “Shared DNA Segments” – the intersection of a segment of my DNA from an Ancestor, with the segment of a Match’s DNA from the same Ancestor. These two segments originate in the same Ancestor and are passed down through two different children of the Ancestor, along different paths to me and my Match. It would be very unusual for me and my Match to wind up getting identical segments. What we “see” in a chromosome browser is the “Shared DNA Segment” – the part that overlaps. Likewise, the Shared DNA Segments with many Matches are different. Since all of these Shared DNA Segments in a TG are compared to my own DNA segment, some of them will start where my segment starts (no earlier), and end where my segment ends (no later).
Now, remembering that each of my segments represented by a TG segment began with a specific Ancestor. Let’s call this Ancestor the “TG Ancestor”. No one ancestral to the TG Ancestor would have the full TG DNA segment (from beginning to end). That full TG DNA segment was created, on one chromosome, by one of the TG Ancestor’s parents through recombination of his/her maternal and paternal chromosomes. So the TG DNA segment has two parts – one from a grandmother and one from a grandfather of the TG Ancestor. Sometimes this can be detected and the TG can be split into two smaller TG, but usually it cannot be readily detected without using somewhat smaller Shared DNA Segments. This may be the same as what you describe in your second sentence.
The words “vertical” and “columns” do not appear in my blogpost; but if you are referring to subdividing a TG, see the previous paragraph. I prefer to not use the term “vertical columns”, because that is too easily conflated with “pile-up” regions.
I encourage you to Trianguate your Shared DNA Segments at one of the companies – it is a simple, but tedious, process – every one of your Shared DNA Segments should go into a maternal TG or a paternal TG or be culled out as a false segment (usually under 15cM). In this process you will begin to understand the many variations – I hesitate to call them “patterns”, because they are actually formed randomly (except the beginning and end of eachTG which aligns with our own DNA segments.
One of the outcomes of segment Triangulation is the idendification of the unique recombination crossover points in our own DNA.
LikeLike
Than you so much for taking the time to explain this in better detail. I appreciate both your caution and the importance of clarity in how these details we observe, are described!
LikeLiked by 1 person
Jim, I wrote this in response to your post:
LikeLike
Jason, I think I’ve figured out this issue. AncestryDNA claims that more than 3 4C can’t share exactly the same DNA segment (the odd are too great). The way this is worded, I think it is pretty much correct – it says exactly the same segment. If they are talking about an ancestral segment, I’ve not seen any two cousins with exactly the same whole segment from an ancestor. What we “Triangulationists” see are overlapping segments – the part of our ancestral segments which overlap – the shared segment is almost always smaller than the full segment. And in a TG, there are virtually no shared (overlapping) pieces of the full TG that are exactly alike, much less the full ancestral segments from whence the overlapping parts came being exactly the same. So it’s trick wording, or trick interpretations, that are being argued. We are all seeing many Matches and segments in our TGs – the numbers are doubling every 14 months. These overlaps are not exactly alike, and the full ancestral segments they came from are not exactly alike. But there are many such overlaps in our TGs. For some reason, some folks are trying to discredit Triangulation. But I’ve got over 90% of my DNA covered by TGs, and virtually every single new segment “fits” easily within an existing TG. And so the TGs grow with more and more Matches. I’m not wringing my hands in disbelief, I’m shouting hurrah – here are more folks who share the same Common Ancestor – and together some of us might be able to sort it out. As new Matches pour in each week, the odds of finding those intermediate Matches increase. By intermediate I mean the 4C to 7C for whom we have a chance of finding a Common Ancestor.
LikeLiked by 1 person
Very good. Very clear. (None of those confusing numbers to deal with. LOL!)
LikeLike
Thanks, Israel – I think this is an important concept for AJ and other endogamous populations. What’s different about A1, A2, etc. is the path they take in passing DNA segments down to me and a Match. I think intermediate cousins, with MRCAs, can help sort it out (to be PART III). These are concepts to use as tools – they are not magic wands to pull rabbits out of hats. There is still hard work to do, but tools can help us whittle away at it.
Jim – http://www.segmentology.org
>
LikeLike
Thanks for making this crucial point so clear.
LikeLike
Thank you. It’s critical that we get past sweeping, broad-brush statements about endogamy, and analyze what is actually happening and figure out ways to deal with it. Stay tuned for PART III.
LikeLike
Karen – I follow Roberta’s blog, and remember this one. She uses “unzipping” in the sense of separating shared segments into paternal and maternal sides. The two chromosomes are not, somehow, tied together. There is no single chromosome with maternal and paternal parts. And all of the IBD shared segments are separate, they just don’t have a label saying which parent they are from. Roberta’s unzipping analogy is about separating shared segments.
LikeLike
I think we are on the same page now. I was referring to the pictorial representations of a chromosome ex: a Chrom browser shows one line for each chromosome but it actually has data for both our paternal and maternal inherited DNA. Just because two people overlap they may not be related.
I have an excellent example but have not been able to include here. It shows my husbands two first cousins one from his maternal side and one from his paternal side if I substituted unknown matches for these cousins we would not know which side of his familial line they received their DNA. Using 1st C demonstrates this really well, ergo unzipping the pictorial chromosome.
Thanks for the nice chat
Karen
LikeLike
Jim I am confused by your opening paragraph. Two of the sentences do not align with my understanding of DNA. Can you help?
“Overlapping IBD segments form Triangulated Groups (TGs).”
I thought overlapping segments did not mean everyone in that group Triangulated. Perhaps only some of my matches who share overlapping segments Triangulate. This would be because some of those in the overlap are from my paternal side and some may be from my maternal when you unzip the chromosome.
“Every Match with a significantly overlapping shared segment will have the same CA!”
Base on the last explanation this statement does not make sense to me either. How can all matches who share overlapping segments automatically be considered to share the same CA. Again if the matches are from opposite sides of my family they cannot share the same CA but they may share an overlapping segment.
LikeLike
Karen, you are correct. I got a little sloppy with the intro. I’ve edited the post. This post was not intended to define a Triangulated Group, or to change what I had previously posted on Triangulation. It takes time to redefine a TG every time I use the term, and to include all the caveats and cautions. The main point of this post was to highlight that there can be intermediate MRCAs in a TG.
I’m not sure what you mean by “unzip the chromosome”. Your paternal and maternal chromosomes are completely separate. You may be thinking about how your paternal chromosome is formed from your father’s two chromosomes…
LikeLike
You might find the article at this link interesting. It addresses the concept of both paternal and maternal DNA shown on a picture of a single chromosome. (Unzipping:)
LikeLike
You all sent me to google to get definition for “elucidate” and “noodle”. These are not in my everyday vocabulary. Guess my age is showing – have a good day.
LikeLike
Pingback: Endogamy PART II | segment-ology
Excellent article! Clear concise writing, well illustrated, you should hire out to edit some other blogs!
LikeLike
Bonnie,
Thanks for the encouragement. I’ve got my hands full with Segmentology – lots more to come.
Jim – http://www.segmentology.org
>
LikeLiked by 1 person
Great post Jim. I wonder what program you are using to make these charts? I would like to make them too.
LikeLike
Peggy
I use Excel and paste in circles, squares and lines – no special program.
Jim – http://www.segmentology.org
>
LikeLike
Hi Jim,
Can you elucidate? I have no experience with making charts like this.
LikeLike
Peggy
Open an Excel spreadsheet. Click on the insert Tab, click on shapes and select a circle then paste it somewhere on the spreadsheet. You can highlight and drag it anywhere. Right click on the circle to change colors or width, etc. Look over all the items you can select any insert to your spreadsheet – noodle around this feature to learn all the possibilities.
Jim – http://www.segmentology.org
>
LikeLike
Thanks again for another great tutorial, Jim. Your post emphasizes why I need to redouble my efforts to persuade my cousins to test.
LikeLike
Thank you, Jim! Your blogs are always so helpful and easy to understand.
LikeLike
Thanks, Nancy
LikeLike