A How-To Example with MyHeritage
This blogpost will walk you through the steps to Triangulate your shared DNA segments at MyHeritage – creating a Triangulated Genome. You can use this same process at 23andMe and roughly the same process at GEDmatch and FTDNA. Disclaimer: This Process takes time (I estimate about 120 hours); but once you are done, you have a powerful tool.
The objective of Triangulating Your Genome is to determine 300-400 Triangulated Groups (TGs), adjacent to each other, covering your 44 to 46 Chromosomes from beginning to end. These TGs group almost all your shared DNA segments, cull out most of the smaller false “shared” segments, and identify recombination/crossover points where your DNA shifts from one Ancestor to another. Each TG will represent one segment of your DNA (equivalent to a phased segment), which came from a specific Ancestor, down a line of descent to one of your parents and then to you. This is segment-ology in a nutshell.
The Process
I will describe this process briefly and then in detail, using MyHeritage as an example.
Very brief version – group segments per the Triangulated Segment icon at MyHeritage (see details below). The Main Steps are basically the same for 23andMe, GEDmatch or FTDNA.
Overview – The Main Steps
1. Download the Segments from MyHeritage
2. Set up the Spreadsheet (headers, set a threshold to cull out small segments, add columns, etc.)
3. Group the Segments (the main, long, process – estimated time: 120 hours)
a. Note any known Shared Match relatives as you go – they help later.
4. Summarize the groups (add a header row and ID# for each group, etc.)
a. Assign partial TG-ID: Chr# plus A-Z (depending on Start location)
b. NB: At this point you should have 300-400 groups. Each group represents one segment of your DNA from a specific Ancestor. This is very powerful information and is useful in its own right. Assignment, in the next step, makes each group even more powerful.
5. Assign groups into TGs – genealogy, logic and/or judgment required
a. Assign each group to a maternal or paternal side.
b. Use Genealogy; logic; “spanning” segments; TG “linkage“; ethnicity, etc.
c. NB: Not all TGs may be determined.
6. Enjoy
In the following sections I will describe each of these 5 steps in more detail.
Section 1 – Download the Segments
Log into MyHeritage and click on the DNA tab.

A menu comes down – click on DNA Matches to get:

Select the kit you want to use – for this example I will use my father’s kit, which I had not Triangulated at MyHeritage before.
Here is my father’s page – note 14,225 DNA Matches. Click on the 3-dot ellipsis on the right.

From the menu select: Export shared DNA segment info for all DNA Matches. Note the little Triangulation icon.

You should get a pop-up with a note that the spreadsheet will be mailed to you in a few minutes:

Section 2 – Set Up the Spreadsheet
Here is, generally, what your spreadsheet will look like:

Notes:
1. Save this CSV file as a spreadsheet. I normally include a date in the title (e.g. JVB MH Segs 20201225) I tend to save various versions during this whole process – as backups.
2. The headers are: DNA Match ID [you’d only need this in case of 2 Matches with the same name]; Name [base person with all the Matches – usually you, but in this case my father, James Vincent Bartlett]; Match name [name of each Match – I reduced the width of this column to hide the names for privacy]; Chromosome; Start Location; End Location; Start RSID; End RSID [these RSIDs identify a SNP – useless info to me]; CentiMorgans; and SNPs.
3. Make a backup copy of this spreadsheet – just in case…
4. Delete the Start RSID and End RSID columns.
5. Move the DNA Match ID column to the far right side [out of your way].
6. Insert two blank columns on the left side; and 10 columns to the right of the SNP column.
7. Divide Start and End Locations by 1,000,000 – changing them from bp to Mbp. This will not change the accuracy of anything (all the digits will remain), but it will make it much easier to analyze and deal with these numbers. [My process for this: create two blank columns next to the Start and End columns – let’s say Start is column F; End is column G; the new columns are H and I; and the first row under the header is 2. Then in cell H2, enter: +F2/1000000. Then copy this cell [drag it] to column I. Check to ensure the numbers in H and I are the same numbers in F and G divided by 1,000,000. Now copy cells H2 and I2 to the bottom of the spreadsheet. Next highlight all these numbers: H2 to the last number in row I. Copy that data (Cntr-C). Move the cursor to F2 and right click on it. From the menu, click on the Paste Special icon with 123 in it (that means copy the result, not the formula). All of the data in columns F and G should now be in Mbp, and columns H and I can be deleted.]
8. Sort the spreadsheet by CentiMorgans.
9. Select a cM threshold, and remove [or physically separate to the bottom of the spreadsheet – out of your way] all segments under your threshold. In my case I used 10cM. This removed about 2/3 of the segments, leaving a group with larger cMs that I would be working with. A lower threshold of 6cM or 8cM will greatly increase the time for this process – not recommended. A higher threshold of 12cM or 15cM or even 20cM will shorten the time, but will leave more gaps. Your choice! By keeping these smaller segments separated at the bottom of the spreadsheet, they are still available in case you want to use some of them later. If you delete them, that’s OK – you can always retrieve them from a backup copy.
10. Type in Header titles*: Hd (for headers); Co (for Company); ID#; Par (for parent initial); Sib (for sibling initial); Ch (for children initial); Czn (for close cousin initial); TG (for Triangulated Group); G2 (for generation 2: parents – the “side”); CA/Remarks (for Common Ancestor or any other remarks you want to save). See the example below. *Each of these will be explained more fully, later.
11. Add in “header” rows for each Chromosome – see example below. This is just a visual separator that helps separate the segments by chromosome.
12. Now sort the “over-10cM” part of the spreadsheet by Chr and Start. Remember to highlight the full rows whenever sorting to keep the data in each row with the correct Match.
Resulting spreadsheet:

Notes:
1. I’ve inserted MH (for MyHeritage) in the Co column. This is not important if you will only use data from one company. Eventually I will add data from GEDmatch, 23andMe, and FTDNA, and I want to be able to sort by company from time to time.
2. There are 2 columns between ID# and Par – these are “extra” and sometimes come in handy when grouping – specifically when one group overlaps another group.
3. The Chromosome header row provides a visual demarcation between chromosomes.
This Chr/Start sort puts all overlapping segments adjacent to each other in the spreadsheet. NB: some of these will be on your paternal side; some will be on your maternal side; and a very few (usually under 15cM) will not Triangulate with either side, indicating they are probably false. Under the next section you will form groups, and each group will be on one side or the other. We cannot know, just by grouping, which side they are on; but we do know all the segments in a group will be on only one side.
Section 3 – Group the Segments
This part is hard work. It’s not difficult – the grouping process is fairly straightforward. But I cannot sugar coat it – it is tedious. I used a stop watch to time each chromosome, and I got better (more Mbps grouped per hour) as I progressed through the Chromosomes. In total it took me 120 hours – a table of hours is at the end of this section.
You can start anywhere. I started at the top of Chr 01 and just worked down (I knew I was going to do the whole thing). In hind sight, you might want to start with a medium or small Chromosome, to develop your “rhythm” – to get your “sea-legs” as we say in the Navy. The two ends of a chromosome are often the hardest – so you may want to just start in the middle of one. Later in this blog post I’ll include some other observations. But for now, just start…
Pick a Match at, or near, the top and put a one in the ID# column. This is the base Match for a while.

I picked “Don…”, the third Match down, because the segment started where the other’s did, but was a little longer. This segment was more likely to Triangulate with others further down the list, whereas the top Match, “vicki”, ending at 5.6Mbp, may not overlap enough with many of the others. It’s really a matter of efficiency. In the end, we are going to “touch” all of the above-threshold segments in the spreadsheet. As you’ll find, it’s easier to get set up with one base Match and “touch” as many as you can, before you have to regroup and start over with a new base Match. NB: Once a Match-segment is “touched”, and added to a group, it does not need to be selected again later. However, use any order that suits you.
So now “Don” is our base Match. Next we search/select Don at MyHeritage (we’ll use his full name, and check the cMs of the shared segment – just in case there there is more than one Match with the same name (I found about a dozen such cases).
This is where two monitors really helps. Put MyHeritage (from the internet) on one screen and your segment spreadsheet on the other screen. From your spreadsheet, copy the Match name cell for “Don…” which has the full name, exactly how MyHeritage has it. Then paste this into the search box at MyHeritage.
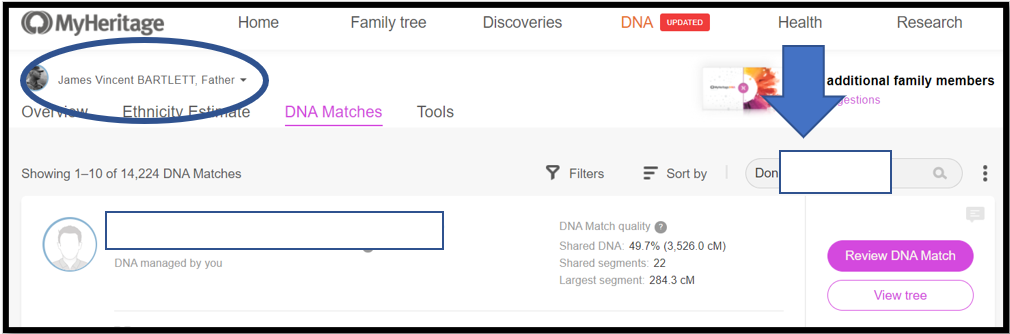
Notes:
1. At My Heritage, my father’s name is circled top left – it’s his genome I am Triangulating
2. Don’s name info is copied into the search box – “Don” is the base Match this time
3. Next: See Downward Arrow and Click on the name; or click on the magnifying glass in the Search box to get a list of Matches per the search criteria. Often the name you want will be the only Match listed. However, sometimes multiple options will be listed and you have to select the correct one (usually the amount of shared DNA will help). Match names like Thomas Jones may return hundreds of Matches – in that case, I punt – and go back to the spreadsheet and select a different Match as the base.
4. Once you’ve selected the correct Match, click on the colored link: Review DNA Match. This will call up a long page focused on that Match. Scroll down past Theories, Smart Matches, Shared ancestral surnames, Shared ancestral places, etc. until you get to the section called: Shared DNA Matches. See below:

Notes:
1. This is the top of the list of 168 Shared DNA Matches between my father and Don. The top two Matches are a close relation to Don and a close relation (me) to my father. Please note the “triangulated segment(s)” icon to the right of these two Matches. In this case Don shares two segments with my father, so I need to click on the icon to insure each Shared DNA Match is sharing the DNA segment at the beginning of Chromosome 1 in the spreadsheet. Scroll down the Shared DNA Matches at MyHeritage, noting which ones share a triangulated segment on Chr 1, and note them with a digit in the ID# column:

Notes:
1. Don and 5 other Matches are shown here as sharing a Triangulated Segment with my father. Indeed, Don matches my father, and each of the Matches also match my father and Don. This is Triangulation in each case; and as a group, they form a Triangulated Group (TG).
2. At the bottom of the first page of Shared DNA Matches (each page has 10 such Matches), keep tapping on “Show more DNA Matches”, until you get to the bottom of the list. The Shared Matches are roughly listed in by total amount of shared cM, so by going to the end of the list you will pick up the shortest Shared Matches. I think this method is the most efficient in “touching” all the Match-segments.
I then picked “Virgi” as the next base Match; copied that name from the spreadsheet cell; pasted it into the search box at MyHeritage; selected the correct “Virgi” from the list of Matches that came up; clicked the “Virgi” Review DNA Match link; scrolled down the resulting pages; and reviewed the Shared DNA Matches between my father and Virgi who had a Triangulated Segment icon. I noted all of them with a “2” in the ID# cell.

Notes:
1. It’s easy to see that the group with ID# 1 and the group with ID#2 overlap the same area on Chr 1 – one of these groups is on my father’s paternal Chr and the other is on his maternal Chr. At this point we cannot tell which is which – more on that in Section 4.
2. I then tried Match “John” and drew a blank – “John” did not have any Shared DNA Matches who had a Triangulated Segment icon for Chr 1.
3. Next I tried “blai” as the base, and “blai” had some icons with Matches who already had a 2, and some new Matches. So we now have:

I now paused to re-sort this part of the spreadsheet – I sorted by ID# and Start. This moved “John” to the top (out of the way – as almost certainly a false segment); and clearly showed the ID#1 and ID#2 groups:

Notes:
1. The color coding in this figure is just to highlight the two groups. I do not do this in actual practice – I let the sorted ID#s speak for the groups.
2. Select the next, ungrouped, Match-segment, “Nanc”, and repeat the process.
3. For ID#, use the numbers 1 to 9 first, then the letters a to z – they will all sort in order.
Grouping Mantra: Select a base – group segments – repeat…
A quick summary of these steps:
1. Select the next base Match in your spreadsheet
2. Copy the Match name
3. Paste the Match name into the search box at MyHeritage – and execute the search
4. Select the Match from the ones listed and click on “Review DNA Match”
5. Scroll down the resulting page to Shared DNA Matches
6. Look for Matches with the Triangulated segments icon
7. Verify it’s the spreadsheet segment of interest (same cMs as in spreadsheet)
8. Type in the ID# character (1 to z) in the spreadsheet
9. Continue down the spreadsheet per steps 6-7-8, until the last Shared DNA Match
10. Repeat from #1.
Notes:
1. Generally pick the next Match in the spreadsheet as the next base. However, if it is a common name, or if it’s a very small segment, or if that Match turns out to share several segments, feel free to select another Match, near the top of the list, as the next base.
– this makes the process go faster
– a skipped Match will often show up as Triangulated later in the process
2. At the end, you can always go back and pick up any “stragglers”. These “stragglers” will usually be smaller segments, which would usually have a very distant Common Ancestor – I’ve actually just abandoned some of these, and moved them to the bottom of the spreadsheet with the under-10cM segments.
3. Be careful with Matches with multiple shared segments – click on the TG icon to insure the TG segment agrees with the area you are working on. TIP: Note the total cM of the Match (with a TG icon) at MyHeritage – if that is the same cM as the Match in your spreadsheet – all is OK – you’re dealing with only one segment and it has to be the one you want. Most Match-segments will be like this.
4. Some groups may be very small, and some may be very large. Many 10-15cM segments in a group may indicate a pile-up which is probably very distant – but at least you’ve grouped these segments to highlight that probability.
5. Don’t worry if the ends of some groups overlap the beginnings of other groups – the company algorithms cannot identify the crossover points precisely – these ends are fuzzy.
6. In my experience, MyHeritage Triangulation has never been incorrect. However, in some cases, segments that should Triangulate are not identified (a false negative), although those segments are almost always identified as Triangulating with other segments in the same group. I think imputation of SNPs is the culprit here. The main point is to put each Match-segment in a group (or cull it out as false).
7. In a few cases the picture got muddled. My best solution is to just skip over this area (these segments) and proceed further down the spreadsheet. Usually the bigger picture becomes clear, and you can return to the problem segments later.
8. Remember the objective of this exercise is to form 300-400 Triangulated Groups, spanning all of your Chromosomes. You do NOT have to adjudicate every shared segment to accomplish the overall goal. The 300-400 TGs are the prize – the important tools that will help you immensely.
Here is a summary of how long it took me to Triangulate each Chromosome:

Notes:
1. I rounded my total of 115.9 hours up to 120 as an estimate for most folks. Hopefully you will have the advantage of using this blogpost to be more efficient than I was.
2. Chromosome 1 was a bear! But I was also working out processes and methods…
3. As noted before, you might want to start with Chromosomes 13 to 22. It’s a good feeling when you reach the end of each Chromosome;>j
Other observations about forming groups:
1. The Chromosome tips (Start and End of each Chromosome) usually have short TGs, and they are sometimes hard to figure out. For a few of these Chromosomes, I actually shifted to the middle of the Chromosome in the spreadsheet, and worked both ways. It doesn’t matter much as every shared segment has to be in one overlapping group or the other (or be false).
2. When searching at MyHeritage for Matches who have the TG icon:
a. If there are only a few Matches (say under 10) in your spreadsheet which could overlap, then just look down the Shared DNA Matches at MyHeritage for the Matches with the TG icon. These few Matches will be easy to find in the spreadsheet – type the ID#.
b. If there are many Matches in the spreadsheet which could overlap, I usually highlight those spreadsheet rows and sort them on Name. This way I can start with the Shared DNA Match name at MyHeritage (with a TG icon) and easily and accurately look down the alphabetized names in the spreadsheet to find the Match name – then type the ID#. [This advice may be more clear after you’ve tried finding some MyHeritage Matches in your spreadsheet.]
c. At MyHeritage, skip over the Matches which show a total cM less than 10cM (or your threshold) – you’ve moved those Matches out of your grouping spreadsheet.
d. Some Matches with a TG icon at MyHeritage may show, say, 18cM, but they aren’t in your spreadsheet. Upon inspection in the browser, the 18cM actually is two segments and the one you’re working on is below the threshold. Aaarrrrg! False alarm. Skip over that Match and continue looking for TG icons.
e. These are my tips – or discover your own “most-efficient” way to find TG Matches at MyHeritage and then find those Matches in your spreadsheet to type the ID#.
3. From time to time, re-sort the Chromosome you are working on. Within a Chromosome, you can sort on ID# and Start location (they sort from 1 to z). This keeps the groups together and quickly shows you the remaining segments to work on.
4. Long segments. Sometimes a long segment will stretch beyond the bulk of the other segments in a group. This usually indicates a closer cousin, with a shared segment that spans across two (or more) segments. This is OK. Later, a segment in an adjacent group may also show a TG with the long segment. I tend to keep the groups separate, because they will probably come from different Common Ancestors down to you, through one closer Ancestor. The long segment is an important “tell tale”, indicating both groups are on the same side, and closely related.
5. Most “tell tales” will come from known close relatives. These are parents, children, siblings, avuncular, close cousins and Theories of Family Relativity (ToFR) that you agree with. Parents, avuncular, cousins and ToFR are particularly useful in identifying the side of a TG. Children and siblings tend to share long segments on one side or the other – usually through several TGs and sometimes over an entire Chromosome. Use the Par, Sib, Ch, and Czn columns in the spreadsheet and type in initials (or other identifier) for these known relatives as you find them as Shared DNA Matches with TG icons at MyHeritage (check the browser to ensure they share on the right segment). I note a Match ToFR in a spreadsheet column for Common Ancestor/remarks. These will help you later.
6. Even with a lower threshold of 10cM, some of the 10-15cM segments don’t always match others as they should. I note that MyHeritage uses imputation to add “most probable” SNPs, in order get larger, more complete segments. But I recall that they don’t use these inputed SNPs when they declare TGs – so even though there is an overlap, sometimes MyHeritage won’t indicate a TG. As a result, not all overlapping segments in a group with show with a MyHeritage TG. I got used to it. After all, every shared segment has to be from one parent or the other (or be false). So if a segment shows as TG with several others, it’s probably true, even if it doesn’t match every segment it “should” match. If you get frustrated with a Match-segment in your spreadsheet, just code it ID# 0. You can ignore it now, or forever. It saves time to “skip over” problem segments.
7. MyHeritage appears to be throttling the Shared Match list after an hour, or so. The “Show more DNA Matches” sticks on the timer and doesn’t show the next list, and/or the next page just hangs up looking for TGs – in either case, you cannot find any more TGs. Options:
a. Click on your browser refresh icon (top left).
b. Use the back arrow, and try a different Match.
c. Turn MyHeritage off; and then back on.
d. Try again 8-12-24 hours later… I was always able to start fresh the next day.
e. Maybe just be content with working on this project an hour a day.
8. Order is not a factor – you can literally work any area of your spreadsheet you want, in any order you want. But the objective is to cover the whole genome eventually.
9. Extra Credit – determine the number of segments: In the beginning, before sorting out the small segments from your spreadsheet, add a column called “Seg”. Then sort the entire spreadsheet by Match name DNA Match ID. Put a 1 (the number one) in the “Seq” column for the whole spreadsheet – most of your Matches, by far, will only share 1 DNA segment with you. Then scroll down the whole spreadsheet and change the 1 to the number of segments each Match shares with you. Yes – this is tedious; but this information (number of segments a Match shares), is very helpful in selecting a base Match, or even understanding that there are multiple segment. However, this is NOT a required step for the overall process.
10. Don’t get discouraged – this whole process gets familiar and easier as you gain experience. Take frequent breaks. The end result is worth it.
Section 4 – Summarize the Groups
When you are done grouping all the segments, take a well earned break! The tedious part is over. This section is easy and a little fun… We are going to insert a header row for each group. We still don’t know which side (maternal or paternal) each group is on, but we do have the Match-segments in groups. I estimate you will get about 300-400 of them.
See the figure below:

In this case, I inserted a new row just above ID#9 in Chromsome 8. I like to use a highlight color. For these “group” rows, add TG in column A; and TG in the Match name column; type the Chromosome number in that column; and the ID# in that column. In the Start Location column type in a number just a little bit smaller than the first segment – I typed in 64.0 over 65.0 for ID#9 and 77.0 over 77.2 in ID#a. The idea is that I can sort the spreadsheet by Chr, ID# and Start and always come back to this list with all the Match-segments sorted in their respective groups.
Repeat for all 300-400 groups. NB: There will always be two groups that start at the beginning of each Chromosome – one for each side. For each of these group headers, use .001 for the Start location – this will sort before any Match-segments and after the Chromosome header (which Starts at 0.0).
Now we can also assign the first three characters to name this groups as TGs.
– The first two characters are the Chromosome number: 01 to 23
– The third character is a letter indicating where the group starts:
A for 0 to 9.9Mbp; B for 10.0 to 19.9Mbp; C for 20.0 to 29.9Mbp; etc.
Here is an example:

In the next section we can start assigning the TGs to sides…
Section 5 – Assign Groups into TGs
This is the final section. After all of the tedium of grouping segments, we now come to assigning these groups to a maternal or paternal side. As mentioned before, each group represents part of your DNA – on one Chromosome or the other. Each group is DNA from your maternal side or your paternal side. The segment data, by itself, doesn’t provide a clue. We cannot assign a side without additional information. We need to use genealogy.
The main method is to use known relatives. If you’ve tested at least one parent, that is gold at this point. You can pretty quickly determine all the shared segments which include that parent, and the remainder would be with the other parent. I used that method to Triangulate my own genome – I have my father’s atDNA, which was a huge advantage. But now, I’m working on Triangulating my father’s genome – his parents, and their generation, are long since gone. So, in this case I have to rely on known relatives. And we can use logic in some cases – more on that later.
There are some workarounds… For instance someone who is half English or half Ashkenazi Jew or half Scandinavian or half Asian or half African… we might be able to tell from the Match names (or Ancestors) in each group which side they were on. It’s a stretch, but perhaps one parent has New England ancestry, and the other parent has Colonial Virginia ancestry… We’d need to really dig into the Trees of many Matches in each group to tease this information out.
I like using Ahnentafel numbers, so I assign the number 2 for the paternal side, and the number 3 for the maternal side. Alternatively, use P for paternal and M for maternal.
In the following example, I have a close paternal cousin of my father – John. Wherever I found John, I could add 2 (for paternal side) to a column I called G2 (for the second generation, which are the parents).

I know that the ID# 9 group he is in must also be paternal (they all share the same Common Acnestor). I can add a 2 to the TG ID and assign that TG ID to each segment in that group:

All of the shared segments in group ID# 9 are now assigned to TG 08G2 and G2 = 2 (paternal side). This 2 on the paternal side is important because, later, I can separate all my segments into paternal and maternal sides, and very easily see how they are adjacent (or not) to each other. They should be adjacent to each other, and this kind of a sort will highlight any gaps.
Next, we will use some logic. TG 08G2 ends in the 76-78 range (discounting close cousin John’s long segment ending at 90.6 – which close cousins often overlap to the next TG). Group ID# a starts at 77.2, which looks like a good “fit” – a little better than the 82 to 96.9 range in Group ID# b. So, I’m going to conclude that ID# a is a continuation on the 2 – side.

Continuing with this logic, group ID# b clearly overlaps ID# a (side 2) by a lot. So it must be on the other side – side 3. So we now have TG 08I3.

We actually have several different ways to make these assignments:
1. Common Ancestors:
– The best way to make assignments is with known close relationships which tell us which side a group is on. Parents, aunt and uncles, and close cousins are strong indicators.
– MyHeritage also offers Theories of Family Relativity (ToFR). My experience with them is mixed – some are clearly correct (they agree with my 46 years of research); some are completely bogus. So, heed the correct ToFRs, and make assignments to a side accordingly. Ignore the bogus ToFRs.
– Also use Common Ancestors with Matches which you work out on your own and are confident about.
2. TG “Fit”: Examine the rough Ends of TGs and see where they fit with the Starts of subsequent groups. All of these groups and TGs have to fit together somehow. None can significantly overlap any others on the same side.
– However, allow close cousins with long shared segments to overlap.
3. Siblings and Children: Not illustrated here are the use of siblings and children. They tend to share very long segments on the same side – spanning several groups or TGs. So, also follow those patterns, which “tend” to stay on the same side from TG to TG.
4. Ethnicity and Geography: As previously discussed, you can also use ethnicity in some cases. You can even make a case for geography – if one side has some unique geographic roots that set it apart from the other side.
5. Other companies & Matches: I should also make the case for Matches from other companies. I’ve done most of my father’s genome, and now I’m reviewing his Matches and known Common Ancestors from FTDNA and GEDmatch and figuring out most of my father’s remaining TGs. The genome we work with (usually our own, but in this case my father’s) is fixed – the recombination crossovers are determined before birth, and don’t change. Although the companies may test a different set of SNPs, the 300 to 400 Triangulated Groups should stay the same. The start and end of each TG may be a little fuzzy, but the bulk of the TG still comes from a specific Ancestor, down a line of descent to one of your parents to you. Don’t worry about the fuzzy ends – think about the thousands of SNPs in a TG coming from an Ancestor.
6. Painting and Clustering: If you use DNA Painter or Clustering and determined a side for some of your Matches, use that information.
Other observations:
1. Double up on all the Chromosome headers – there needs to be two for each Chromosome – one paternal (put a 2 in the G2 column) and one maternal (put a 3 in the G2 column).
2. Something I thought about, but haven’t tried, yet. Set the threshold at 20cM and Triangulate those segments fairly quickly (you would skip over a lot of Matches with TG icons who shared less than 20cM). Then re-sort the full spreadsheet and set the threshold at 15cM – then re-sort the 15cM+ spreadsheet by Chr and Start. This has all the segments in order, but the paternal and maternal sides are comingled with the new 15-19.9cM segments. These new segments should be easy to Triangulate other segments already identified with TG IDs. I predict some of the larger TGs will be subdivided in this process. Then drop the threshold to 10cM and repeat. That would take 3 passes through the whole genome, but it may go faster than by starting with a 10cM threshold. Maybe I’ll try my brother’s MyHeritage kit that way…
Section 6 – Enjoy
This in a MAJOR MILESTONE in Genetic Genealogy. Now that you’ve put in the work to create 300-400 TGs that cover your genome, enjoy the benefits of this great tool!
1. All new shared segments (from any company) should quickly and easily fit into an existing TG. You have the framework for organizing all your Match-segment data.
2. Each TG comes from a Common Ancestor – even ones behind a brick wall… Review the blogpost: Let the Matches Tell Us the Cluster Common Ancestor here. TGs, just like Clusters, are groups focused on a Common Ancestor, and the Matches in a TG may be able to tell you who the Common Ancestor is.
3. When you know which line a TG is from, tell your Matches! Focus on your Matches’ Trees to find that line. With that focus, I have found a collateral line in a Match’s Tree and built it back to our Common Ancestor – many times.
4. Walk the Ancestor Back within each TG. See here.
5. Pile-on (not pile-up). In several TGs I’ve gone back to the Matches asking if they had a particular Ancestor in their Tree. Even when they had no Tree, they sometimes confirmed that Ancestor I was focused on. I’ve collected dozens of Matches who all share the same Common Ancestor with me. I’ve done this with a growing number of different TGs. This is very strong evidence that the DNA segment (represented by the TG) is from that Common Ancestor.
6. Play with these TGs. You can easily use DNA Painter to paint the TGs! All the Matches in a TG could also be painted if you want. Think about Painting virtually every one of your Matches…
7. You can easily use the G2 column to sort your TGs into a paternal side and a maternal side.
8. As mentioned before, add the data from other companies. Consolidate all your Matches and segments into one comprehensive spreadsheet.
The End of Triangulating Your Genome
I’ve enjoyed putting this blogpost together, and sharing some of my concepts on Triangulation. If you try this, please post some comments about your experience and/or any suggestions for improvement.
[10A] Segment-ology: Triangulating Your Genome; by Jim Bartlett 20201229

Pingback: Why Don’t the Segments Triangulate? – Family Locket
Pingback: Is This a False Segment? – Family Locket
Jim,
Thank you for this excellent blog. I am part way through the long process of grouping, using My Heritage as in your method. I’m a beginner with this, so please could you clarify something for me.
On chromosome 10 I have a match with CW who I know is a 3rd cousin on my mother’s side and I know our common ancestors. Working on your method, I find that CW triangulates with PF on ch 10. So PF must be on my mother’s side and must have the same common ancestors as CW and myself. OK so far?
Now I’m on chromosome 14. There’s no match to CW, but there is a match to PF. When I look at PF, he triangulates with AB on ch 14.
So can I say that CW, PF and also AB all share the same common ancestors with me, and AB is also on my mother’s side? If that’s correct can I repeat the process, finding AB on some other chromosome, looking for triangulations, then those also have the same common ancestors? Or am I missing something?
Thanks
Dennis
LikeLike
Dennis – we cannot “chain” Ancestors like that. Each segment is independent. I have lots of Matches who are multiple cousins with me on different lines. It is fair to treat a Match relationship as a “clue” across multiple segments; but not as a hard fact. I am not satisfied until I can find multiple MRCAs in a TG along the same line.
LikeLike
Jim,
Thanks for the explanation. I’ll keep working on it.
Dennis
LikeLiked by 1 person
I did some additional tasks. I add a column with number consecutively 1 on. then if I want to sort and delete rows if I ever want to get back to the original order I can. I used a pivot table to get total number of segments and total cm. I could have also done largest and smallest segment for a match where there was more than one segment. I then went to the original file and used a lookup and added those columns to the data. Lastly I color coded the matches based on cm. I think left all the matches in. I did not delete any. I went to the ones with color over 20cm and then could see those that overlapped that were smaller. The colors helped me to find the higher cm matches to work on. I could also just look at those that only had one segment matching because of the added column I did. Rather than deleting or moving low matches I would hide them if I wanted to. Then could easily unhide them later.
LikeLike
RH, Thanks for your feedback, and suggestions. My spreadsheet is over 20,000 rows now, so I move the segments that don’t Triangulate to sperate spreadsheet – they are almost all under 10cM, probably very distant, and in general not worth my time. I color code the segment data blue and pink, to clearly identify paternal and maternal. I also have a column for side (2 is paternal and 3 is maternal), and most of the time my spreadsheet is sorted by side + Chr + Start (Mbp) – so all segments are grouped into TGs. As new, promising, Matches are added to a TG, I review the whole TG for the Most Distant Common Ancestor. Jim
LikeLike
I wish you would create a blog for the other websites. I share this one with dna matches of 23andme and familytreedna and gedmatch. But there are some differences for each website and some users need more hand holding. It would save me some time. So if you do not have anything else to say sometime that might be an idea. Also perhaps also make a document because each website changes and if you had a document you could edit it to update. Anyway just an idea.
LikeLiked by 1 person
RH,
I plan to do blogposts for the other companies, but not just yet. Each one is more complex, and requires some judgment. I’ve got several other posts brewing right now… Jim
LikeLike
Jim, in Section 2, item number 10, you said you would give a more detailed explanation of how you label the header rows later. Have you done so, and if so, could you please add a link to that explanation? Thanks!
LikeLike
Emily – I meant later in the post. See the yellow bars later in the post. There are header rows for each Chromosome and header rows for each TG. Jim
LikeLike
Pingback: AutoSegment Triangulation Cluster Tool at GEDmatch – Open Source Biology & Genetics Interest Group
Pingback: AutoSegment Triangulation Cluster Tool at GEDmatch - Search My Tribe News
Pingback: AutoSegment Triangulation Cluster Tool at GEDmatch | DNAeXplained – Genetic Genealogy
Jim,
Using data from MyHeritage, I have just finished identification of five TG’s on Chromosome 16. Previously, I worked Chromosomes 17-22. My approach is to do pairwise comparisons between all shared matches >15cM on each chromosome. On Chromosome 16, I did 780 pairwise comparisons. That’s a lot of comparisons, but I’ve found a way to do them fairly quickly and efficiently. Then I use Autocluster at Genetic Affairs to form triangulation groups. The data are inherently “noisy,” and the auto cluster routine really helps to see the big picture in a matter of seconds.
TG 16F2 has 18 matches. Inspection of the Excel data sheet shows that this TG starts at about 54.0 Mbp and ends at about 82.0 Mbp. TG 16G2, which has 12 matches, starts at about 64.0 and also ends about 82.0.
There is significant overlap between TG’s. 16 of the 18 members of TG 16F2 extend deep into TG 16G2. Just two members of TG 16F2 end at 64.0, which would actually line up well with the start of TG 16G2, _IF_ 64.0 could be considered the boundary between 16F2 and 16G2.
The autocluster plot shows several dozen gray cells outside the boundaries of 16F2 and 16G2 that appear to link these two TG’s into a larger 16F2 / 16G2 “supergroup.” This suggests that both groups are related via a deeper, common ancestor.
I’m confident both TG’s are on the paternal side. That’s because there likely was little or no admixture between the ancestors of my father who was born in Georgia (USA) and my mother who was born in Germany, having lived there until age 19 at the end of WWII. At MyHeritage, I do see glimpses of DNA that appear to be from my mother’s ancestors. However, the preponderance of my matches appear to be from my father’s side, and the relatively large group sizes argue for that.
Aren’t triangulated groups supposed to match up heel-to-toe along the length of a chromosome? Triangulation Groups 16F2 and 16G2 overlap to a significant degree, which is confusing to me. I’m not sure what to make of it.
Where should I draw the line between these two triangulation groups, 16F2 and 16G2? What might be going on here?
LikeLike
Andy,
I have no hard answer. I think your two TGs are actually one TG from a close Ancestor. But I, too, keep them separate – they are probably from a husband/wife couple and one TG goes back on one and the other TG goes back on the other. One of your Ancestors (a child of the couple) passed both TGs down as a larger segment. Remember your father passed down an entire Chr 01 to you that had a lot of embedded segments from more distant Ancestors – I’m working on a blog post to more fully explain this. Jim
LikeLike
Hi, Jim. I’ve been dabbling in your methodology hoping it will help sort out my endogamous lines and have a couple of questions. First, in the scenario where a) Match A triangulates with Match B and others but not Match C; b) I next go to Match C and immediately find a triangulation with Match B. Can I assume they’re all one group and therefore stop going down the Match C shared matches list (assuming there are not a lot of other unaccounted overlapping matches that could be picked up on that list)? My second question is: When I’m close to the bottom of a shared matches list and MH starts acting up, can I just use the Chromosome Browser to check any other triangulations amongst a few remaining matches who have overlapping segments?
LikeLike
terrhart,
Q1 – mostly yes – you can assume that Match C is part of the AB Triangulated Group; you also can stop going down the Match C list; but you may miss some unaccounted Matches that would be in the TG. Generally, if you go down the list of all “un-TG’d” Matches, you’ll catch any “strays’.
Q2 – again some yes a little no – a browser can only show overlap, but cannot guarantee Triangulation. However, if the Match only shares one DNA segment with you (and vice versa) – it’s usually very safe to assume it’s in the same TG you’re working on. And by extension, if there are multiple segments and one falls in the same location you are working on, it would be safe to assume Triangulation with that overlapping segment.
In general, by going down the list of Matches (on a chromosome; in a spreadsheet), I see each one at least twice – I like this as a sort of a Quality Control check.
Please let me know your experience with TGs and endogamy. We need that kind of feedback from multiple sources. Jim
LikeLike
Thanks for your reply, Jim. Your method does hold a lot of promise if I can only work my way through it. Due to the endogamy, there are a lot of shared matches and to get through to the bottom of the list for just one match, there is a lot of stopping and starting (MH issues can mean hours for just one match if it’s a long list). On your response to my second question, if I use the chromosome browser at MH on those few matches in an overlapping group who are “left over,” the browser shows when they do triangulate with a bracket (I have the overlap set at 8 cM). I’m therefore a bit confused on your caution there. I would have thought the chromosome browser triangulation feature would be drawing from the same info as the triangle icon. Can you explain further?
LikeLike
terrhart, For Triangulation we need you and A; you and B; AND A and B to all share overlapping segments. In a browser you can see you and A and you and B; but we cannot see A and B – except in a browser that also shows Triangulation. If you can call up A and B in your browser AND get the Triangulation grouping around the segments you have Triangulation. So, YES, if you see the Triangulation “loop” around the segments.
LikeLike
Thanks, Jim. Yes, that is what I was referring to – the triangulation bracket/loop. It seems for me, with all of the endogamy, when there are only a few remaining matches in a group with overlapping segments, it would be much more efficient to check for triangulation using the chromosome browser than scrolling down through the extensive shared match lists of those 2-3 remaining matches in an apparent group. I haven’t been tracking my hours very diligently but it appears this will take a longer than it took in your case. Thanks again for your help.
LikeLike
terrhart, I must confess that I tried several different methods – it’s a long, boring process and I tried several methods. Each time I tried a different method, it too slowed down, and I reverted or tried other ways. I’m not confident that there is one best method – other than just working through each Chromosome. I couldn’t find a consistently efficient method… But the end product is a powerful tool. Jim
LikeLike
While I have way too few matches that I can assign to positions on my tree, and I haven’t finished assigning matched segments to triangulation groups, I have reassigned one segment from an MRCA with the segment owner to a more distant CA on another line based on a shared segment on DNAPainter. This on my father’s, predominantly Jersey Dutch, side; doing the same on my mother’s, predominantly 19th century Scotch Irish immigrant, side will be much more challenging.
LikeLike
Richard,
One of the important features of Triangulation is when you can determine the side. I have had a number of Matches with Common Ancestors, only to find that the shared DNA segment is on the other side – the DNA came from my mother and the Common Ancestor is only on my father’s side. We are still genealogy cousins on my father’s side, but there must be another Common Ancestor on my mother’s side who passed down the shared DNA segment to both of us. As you “Walk the Ancestors Back”, the TGs become more and more like laser pointers. Very powerful tools. Jim
LikeLike
That’s what I’ve generally been doing, but this Smith is in a richly populated zone of chromosome 3 (over 100 matches assigned to TG 5, with a lesser but still sizeable number assigned to TG6) so what I’ve been doing is going through matches until I find a match to one of the established TGs, then going to the next unassigned match. Ms. Smith hasn’t shown up among the top matches yet, so I’m going to leave her until I’ve finished with this zone, then do a more in-depth search for her.
Is there some way to pull the match information out of DNAGedcom?
On a side note, on a previous segment of chromosome 3 I found a large number of Holland Dutch, triangulating with one of my Theory of Family Relativity matches. The MCRA for the theory was our respective 4G Grandparents on one of my Jersey Dutch lines. Considering that most if not all of my Jersey Dutch ancestors were New Netherlanders prior to the English takeover in 1664, these folks are most likely 10th cousins or more distant.
LikeLike
Jim…. oops, I forgot to include the numbers”
Doug and Jacob:
Half-identical segment
Physical Position (bp): 92379081-140709905
Genetic distance: 39.52 cM
Number of SNPs: 8123
Doug and Jacob:
Half-identical segment
Physical Position (bp): 140961792-170075171
Genetic distance: 29.24 cM
Number of SNPs: 4992
Doug and Susan:
Half-identical segment
Physical Position (bp): 121573903-158867973
Genetic distance: 33.88 cM
Number of SNPs: 6117
LikeLike
Hi, Jim,
When last I posted, I was working on MyHeritage, but I just felt bogged down with my 19500+ matches there and I was trying to find something about 2-3 matches that are only on 23andMe, so I thought I’d give this a shot there. That went good for a couple of weeks until things started looking suspicious in the Relatives in Common function, After a little analysis, I opened a problem report with 23andMe and I think they’ve removed the problem they introduced with some recent changes now and things appear to be working correctly again.
Now, to why I’m writing. I looked have run across an issue where a number of have a break in a segment on chromosome 4. The break is only 0.26Mb as the first section ends at 140.709905 and then resumes at 140.961792. That seems to me to be a glitch of some kind rather that a crossover. Also, I just found another match that doesn’t have the break. I’m including the browser data below.
Would you recommend treating these as separate triangulation segments, or since I have this bridging segment, and the break is so small treat them as a common segment?
Thanks as always for you guidance and opinion.
Doug
LikeLike
Doug, a few thoughts:
1. I, too, have well over 10,000 Matches at MyHeritage – that’s why I culled out the under 10cM ones, it reduces a lot of the harder-to-work-with small segments. After you’ve formed TGs with the larger segments you can always add the small ones back in – they are then easier to fit into existing TGS.
2. I’ve heard that 23andMe has changed the meaning of their “Yes” column in the past few weeks – I haven’t been back to check, but a word of caution now.
3. There are often some small breaks in our raw DNA data – think of a diamond – they all have some imperfections. If you find a lot of them at one spot, it’s probably a flaw in your DNA. I have three small areas where I do not match my father as I should – I make a small segment out of these three holes and put them in my spreadsheet to remind me. If I have Match-segments on either side, I just count it as one full segment. Almost all of the commercial algorithms gloss over minor data glitches – so I do too. As addition data is added, it usually sorts out. Jim
LikeLike
Jim, thanks for your response.
1) I did cull at MH and then as I was working through some of the matches of some that I knew personally, I added some of the smaller segments back in to the spreadsheet.
2) Yes, I saw the write-up in DNAeXplained-Genetic Genealogy and forwarded her this response from 23andMe Support, that they had sent me in response to my complaint and request for escalation:
” Hello Doug,
Thank you for your reply. Your request has been escalated to me for review. I understand that you have noticed changes as to what “Yes” means for DNA overlap. We are aware of this change, and are looking to resolve this now, so that “Yes” indicates the presence of the same shared DNA segment between all three relatives. The reversion to our previous segment matching logic should be reflected within your account soon – I appreciate your patience in the meantime.
Best,
Louise
The 23andMe Team ”
Based on the fact that they changed none of their tutorials about using the Shared DNA (it still speaks of the “Shared DNA” column) and the wording of this update from Louise, I don’t believe these were intended changes, but bugs introduced by programmers, who had no insight into the original code and its purpose, when making changes that weren’t supposed change anything functional, just descriptive (one of the biggest problems of undocumented or poorly documented code – it makes maintenance a b**ch – 🙂 I don’t have anything saying this is actually what happened except experience gathered over 40+ years in the computer industry.).
3) Yes, you describe this kind of like what I imagined, but I thought of the flaw being in the DNA of one, so far unknown ancestor, who had a flaw in his/her DNA that his sibling(s) didn’t have. So, some of my matches who come from my MRCA have the flaw while other matches, without the flaw, descend from her/his sibling(s) who didn’t share it. Doug.
LikeLike
Hello, Jim. I’m making my second attempt at following your system. Still on chromosome 1.
The first segment on chromosome 1, from 0.75Mbp to 57.23Mbp, belongs to a paternal first cousin. Up to 9.3Mbp, it matches no one on MyHeritage except one of his daughters, who shares that entire segment. Another daughter shares from 15.68Mbp to the end of the segment, and 15.68 falls right in the middle of a group of matches from roughly 9.3Mbp to 22 Mbp.
Question one: Should I assign my cousin’s segment from 0.75 to 9.3 to a separate TG?
Question two: I should copy my cousin’s segment to each succeeding TG that triangulates with it, correct?
Question three: I should ignore the breakpoint at 15.68Mbp, as that reflects his daughter’s crossover points, not mine, correct?
I have one match at 17.4Mbp to 22.17Mbp that overlaps a couple of triangulation groups, but doesn’t triangulate with either of them. He had some triangulations, but all to segments under10cM (my cutoff.)
Question four: I should treat the above described segment as a false segment, correct?
LikeLike
Richard – good questions:
one- no – assigned *every* segment to the TG(s) it matches/Triangulates with – follow the data. You only have two of each chromosome, so all true (IBD) segments must fall into a segment on on chromosome or the other.
two – yes – that’s what I do. Alternatively you can just remember when a close cousins segment straddles two or more TGs
three – yes – by that I mean a Match doesn’t dictate your crossover points. But a lot of data points will.
four – yes – I leave them in the spreadsheet until I’ve Triangulated all the other overlapping segments, and remove them after that.
The long chromosomes are the hardest, and the TGs at the tips are also hard. If you get bogged down, Try, say, Chr 09 and start in the middle:>j
LikeLike
My problem the first time was using a first cousin as my initial base. Tip: don’t use close relatives as a base, they have long segments that span multiple TGs, and a lot of them, meaning you’ll be going back and forth finding matches on segments you’re not working on a LOT.
Tips specific to MyHeritage: if you look at the shared cM for both you and your base with a match before hitting the triangulation traffic light, skip it if either one is under your cutoff point. After hitting the traffic light, if the longest segment of the match is under your cutoff, don’t bother trying to find them in your spreadsheet. You can find a match in the spreadsheet faster if you mouse over the matching segment and see where it starts.
LikeLike
Richard – good tips! I rarely use a close Match as the base, but not when a close Match spans several TGs. The CA with the close Match should apply as a laser pointer in all of the TGs the close Match covers, Jim
LikeLike
How do you use the MyHeritage ID for finding a match who is difficult to find with a name search? 42 pages to find a certain Smith is a bit much.
LikeLike
Richard,
I know what you mean – been there, struggled with it. I now use reverse logic to find a SMITH Match. Each Match will be in one of two TGs (maternal side or paternal side). When I work down my list to a SMITH, I skip over it and work on the other Matches (with overlapping segments). The SMITH will show up as one of the Matches in a TG, and then I’ll have him or her. The SMITH will then be in a TG and I don’t even need to look it up. Actually, working down the spreadsheet has the benefit that many Matches will TG with the first base Match – you don’t need to look those Matches up again – they are already grouped. When you finish with all the Matches with the first base Match, you then jump to the next un-TG’d Match as the new base and work down – grouping all the Matches with the new base. Some of these will be repeats (which you’ve already grouped) and some will be new ones, added to the TG. At some point a new base Match will only TG with new Matches and you’ll have started putting together a new group. Many of your Matches will never be a base Match – force the SMITH Match into this category, by skipping over it. Jim
LikeLike
My confusion is in knowing who gets numbered together. Do they all have to have the same start and end and cMs? Or do they just have to triangulation in same chromosome?
LikeLike
James,
Thanks for your questions. I have not done any videos, and instead try to outline information about segments in this blog. I’m trying to provide directions for regular genealogists, but sometimes miss my mark. Segment Triangulation is a complex concept. Actually the concept should be fairly simple: You and Match A share a DNA segment. You and Match B share a DNA segment that mostly overlaps the segment you share with Match A. Then, IF Match A shares an overlapping segment with Match B (overlapping the segment you share with A and B), we have segment Triangulation. Each segment is a long string of markers (SNPs) – usually 1,000 or more. Think of this long string as a unique password. No other string of DNA at the same location will have this same string of markers – the same password – except for people who got that string from the same Ancestor. [in theory, there are exceptions to this “rule” – very low probability, like you may be struck by lightning on a sunny day]. Just go with the premise – it works for genealogy, and more importantly for Triangulation.
So this should answer your first question – same start and end? NO. In fact most folks will get different segments from a Common Ancestor. What we “see” in a shared DNA segment is only the part we share. And the 3 segments involved in a Triangulation are usually different – the key factor is that they must overlap each other enough the be proclaimed a *match* by the companies (whose algorithms are comparing all the markers). Such a 3-way match can only occur on one chromosome. The second key factor here, is that such a 3-way match *can only occur on one of your two chromosomes* – your maternal *or* your paternal chromosome. As such it has to come from one of your Ancestors, down a line of descent to one of your parents, to you. And the same holds for A and B. NB: if you got this segment from your father’s side, it could be from either side for A or B. In other words, the Common Ancestor could be either paternal or maternal for you or A or B. In general, you have to compare your Tree to their Trees and determine who the CA is and on which side it is for you.
In most cases, we build our Triangulated Groups up with more and more Match-segments. If you had 3,700 Matches and 370 TGs – you’d average about 10 Matches per TG. Among those Matches in each TG, we always hope for a close known relative – parent, aunt/uncle, 1C, 2C, 3C – because this usually tells us which side (for us) the TG is on. This lets us build the Chromosome Map.
Back to your original confusion – number together the overlapping shared segments that MyHeritage says are Triangulated. If a Match has more than one shared segment with you, make sure you only number together the one that overlaps the others in your spreadsheet (on same Chr).
Revisiting the same start and end. In your body – your DNA – you have fixed start and end points for segments of each Chromosome from different Ancestors (these are recombination crossover points). So the segments you share with your Matches are limited by those two points, and *some* of your shared segments with Matches will start or end at those points, but not necessarily. When you “fill up” a Triangulated Group with many shared segments, the start and end points of the TG will tend to be the start and end points of the DNA segment from your Ancestor – but remember the DNA data we get as genealogists is fuzzy (not exact), so don’t expect all shared segments to precisely line up.
Hope this helps, Jim
LikeLike
I’m still a little uncertain about the triangulation of the Genome into groups at My Heritage, so I’ve broken down my theoretical situation as simply as I can. My question is the following, if:
Match A on Chr 1, starts at 1,000 on the segment, and it ends at 2,000;
and Match B on Chr 1, starts at point 1,500 and ends at point 2,500 – and MH says A & B triangulate;
then, Match C starts at point 2,125 ending at 3,200;
so Match C triangulates with Match B, but not with Match A (because A does not overlap with C);
does ABC all triangulate on the same side?
I realize that if a Match D, starts at 1,500 and ends at 2,500 and triangulates with all, that would give me my answer, but that doesn’t always happen.
LikeLike
Linda, Yes! I probably need a separate blog post about this – maybe “Chaining Segments in a TG”. Yes, if U=A, U=B and A=B then they form a TG. From then on anyone* that U match, who also TGs with a segment in the TG is added to the TG (now you can even add parents, siblings, etc). When you have a TG with 20 Match-segments, it is not uncommon for some of the Matches to not Triangulate with others. Sometimes this “chaining” can really stretch out a TG.
NB: anyone* really means an overlapping Match-segment (we have to be careful when a Match shares more than one segment with us, that we are referring to the overlapping segment).
The key is that every shared DNA segment has to go into a TG on one side or the other, or be false (usually under about 18cM at MH). If you are in doubt about a segment, skip over it, and Triangulate the rest. Then when you come back to the “problem” segment(s), it will be much clearer what the two options are. The “problem” segment should TG with some segments in one group and NOT TG with any segments in the other group (on the other “side”).
Jim
LikeLike
Thank you Jim, I’ve been working on my Genome for three months and that question has been top of mind. This really helps. And thanks for getting back to me. I so appreciate your work.
LikeLiked by 1 person
So in MyHeritage, the chromosome browser for each chromosome is pictured as having 2 halves. Do I group all people that have triangulation in the same half or only group people that have triangulation in same part of half? Sorry about this. I want to make sure I’m doing it correctly. I wish I could include a picture. I will do my best to explain my confusion. This is an example, not a replica of a chromosome. Let’s say chromosome 1 has 20 spots on left side and 20 spots on right side. Myself and A match on spots 2-5 on left side. Myself and B match on spots 3-6. A and B match on spot 4. So all three of us triangulate? Is yes, then do I continue to find matches that triangulate for me and A on spots 2-6 or do I start a new TG? I hope that makes sense and I apologize for not getting it right away.
LikeLike
James, the chromosome browser at MH shows one of each chromosome (as do all the other companies). For instance, that is your chromosome 1 – it’s really just a bar to show, graphically where the shared segments are. The shared segments which are shown may be on your paternal Chr 1 or your maternal Chr 1 – they are co-mingled. And that’s one of the big things we are trying to sort out by forming Triangulated Groups. I think what you are seeing are the two colors from the two Matches you are looking at. Note the color under each match – it corresponds to the color of the shared segment bar. On the upper right of the browser is a link to Add or delete DNA Match – try it. You can add any Match and they will show up in a new color on a third bar of each chromosome.
So you’re looking for the segments that overlap – actually you’re looking for segments that have an oblong “circle” around them – when encirclement means those segments are Triangulated. Those are the ones you want to group. Jim
LikeLike
Thank you Jim. I’m going to continue putting the numbers beside what I guess should be in the same numbered group. Just seems strange that there would be more than 20 people in one group.
LikeLike
James,
Using this method you should get about 350-400 groups – so divide the number of your MH Matches by 400 to get an average – some will be larger, and some will be smaller. Jim
LikeLike
Greetings Jim. I am very confused. Is there a video or directions for dummies?
LikeLike
Thank you for this excellent lesson on Triangulation. I’m well into my 120 hours. I have endogamy in my family. Grandfather married cousin once removed. I find that on a single chromosome a person is in two TGs. Do I leave them in two separate TGs? Thanks. Arden
LikeLike
Arden, It’s up to you. The data remains the same. I keep the two segments, and usually find that they are from the same close ancestor (grandparent, Ggrandparent, for instance) and then they split into the two parents going back. This means the person in two [adjacent] TGs is almost surely a close cousin (before the split). Jim
LikeLike
On My Heritage, I have six matches on the same chromosome with the same start and stop numbers. They all match me at 30 cM. I put these matches onto the Chromosome browser; only half are fully triangulated, The other half are half triangulated, yet they all had the same start and stop numbers. Am I reading this wrong?
LikeLike
I’ve found similar on several matches. Also, you can get different results (no triangulating segment in common vs TS in common) depending on whether you use the Chromosome browser stand alone tool or go through the DNA match page. For nowI don’t trust any MyHeritage triangulated segments and instead am doing the analysis with GEDmatch. Genetic Affairs will make the AutoSegment cluster spreadsheet for all four DNA sources: 23andme, Family Tree DNA, GEDmatch, and My Heritage. If a MyHeritage segment overlaps with a GEDmatch TS, then I will look at it.
LikeLiked by 1 person
In this case, all these matches are on MyHeritage and I’d like to understand why in the triangulated segment numbers, say 0-10, my six matches all have the same numbers. When I go to the Chromosome Browser, one to many, and pull up these same matches, they all triangulate, but the triangulated brackets are reduced in size for three of these matches, which reduces the visual size of the match for all. Again, they all match at 30 cM at the start of Chr 15.
LikeLike
Linda,
The reason segments show Triangulation shorter than the overlap is due to imputation. MH imputes some SNPs – that is they add in for missing SNPs (using the most likely value of the SNP at that location). This tends to make shared DNA segments longer. However, it is my understanding that, MH uses the shorter segments (only the actual tested values) when they do Triangulation. Thus some of the Triangulations are shorter. Jim
LikeLike
Watchinit,
I have been doing segment Triangulation since 2012; and I find the MyHeritage Triangulation Icon to be very accurate. That is to say the Triangulated segments are on the same chromosome and overlap each other, and Match each other. I do note that the end points are often fuzzy (due to imputation and their process), but the segments can always be relied upon to be on the same side, from the same Ancestors (say, over 99% of the time). Always remember we have two of each chromosome, and segments which overlap in a browser may be on either chromosome – when they Triangulate, they are on the same chromosome, and come from the same Ancestor. This is more accurate than Clustering, and can be much more comprehensive (i.e. – all segments can be Triangulated, or culled out as false.)
LikeLike
Linda, I’m not sure what “half Triangulated” means – to me me and two Matches either Triangulate or we don not. Clearly I share a DNA segment with every one of my DNA Matches – the key point is when these segments overlap, do they also share with each other (forming a Triangulated Group on one chromosome). The browser does not (can not) tell you which chromosome the shared segments are on. When they Triangulate, we know the Triangulated Matches are on one Chromosome – but we still don’t know which one (without some genealogy hints). So, in this respect, Triangulated Groups are like Clusters – they are from one Ancestors, but, without more genealogy input, we don’t know which side (mom or dad) they are on.
LikeLike
Hi Jim, I read your blog the other day and was fascinated. I am new to genetic genealogy; did my first autosomal test just about a year ago. Recently I have been working on visual phasing using the Stephen Fox method. But when I read your blog I decided to take a break from phasing to try your method and see how they compare (and hopefully agree!).
I got my MH data and loaded the spreadsheet yesterday. I was going to follow your suggestion and start with one of the shorter chromosomes. But when I opened my mail this morning there was a request from a match asking if I could help find a CA. I looked at her match; single segment of 41.3 cMs at the beginning of Chr 15 and thought, why not start there?
Well, I’m not sure what I have gotten into. Maybe I’ve done something wrong and you can point it out. Here’s what transpired.
Following your blog I looked for the longest segment in the block starting at the beginning of 15. Found one that was 57.1 cMs (20.0 to 45.1) and following your blog started building group 1. There were 55 matches who triangulated with this person. Then I moved to the next entry and started group 2. There were 8 matches that triangulated but two of them had already been placed in group 1. I had no idea what this meant but thought I would just continue on.
Group 3 resulted in 10 matches; 3 that were already in group 1, five already in group 2. To make a long story a little shorter, I identified 12 groups. Only the 6th one was ‘independent’, i.e. not already in another group.
All in all there were 144 matches in this segment area. No overlap with other segments. 52 of them did not triangulate, including the match that started it. Could a 41cM segment be invalid?
Before I continue on I just thought I’d ask if anything jumps out at you.
Thanks, Herb Swain
LikeLike
Herb,
It’s good to see you working on segment Triangulation. As you know the DNA is random, so it often throws us a curve ball. The key to remember is that we only have two Chr 15s – one on each side – and virtually never will a segment from one side Triangulate with any segment on the other side. When you start with a 57cM segment, it probably spans several TGs. So the trick there is to look at the list of 55 segments and see if there isn’t some natural break (there usually is). In a Chr+Start sort, this would be a place where say 20 of the Match-segments pretty much TG with each other, and the other 35 form their own groups, with the 57cM segment spanning both. This is why it’s hard to automate this – some of your Matches will be close (2C-4C) and most will be more distant. When you think about it, you get one long segment from your father called Chr 15. If you could break that down, it would probably be two segments – one from each of your father’s parents. In the next generation, the probability is that one of those two (grandparent) segments would be subdivided by the Great Grandparents – meaning you’d have DNA (on Chr 15) from only 3 of your Great Grandparents – and one of your grandparents may have passed two segments as one.
So look down your Chr 15 spreadsheet and see if you can detect two (or three) groups within the 55 Matches. Let me know… Jim
LikeLike
Just to point out that the start of C15 has a pileup region from 10-20 (according to the DNA Painter settings)
LikeLike
Graham Hart, none of my segments begin before 20.0
Jim,
Everything you say makes sense so I sorted and started looking for the patterns but nothing stood out. Yes, I did notice some major groupings, e.g. 10 matches starting at 31.1 and 10 at 35.1, but still no patterns between groups jumped out. So I decided to reexamine the triangulated matches to ensure I hadn’t fouled up.
I discovered an interesting point. The Start for a segment involved in a match is not necessarily the start point of the triangulation. That is, Match A segment start may be 20.0 and stop at say 35.0. But Match B segment may start at 26.5 and extend to say 40. So the triangulated segment is from 26.5 to 35.0 as shown by MyHeritage chromosome browser.
This would indicate to me that to find the patterns I would need to add two more columns to the spreadsheet; Triangulated Start and Stop.
Another anomaly stood out that didn’t make sense. In one case Match A segment is 20.0 to 35.6 and Match B is 27.8 to 46.7 but the browser reports the triangulated segment as 31.1 to 35.6. Why wouldn’t the triangulation start be 27.8?
I’m going to continue on to create those new columns just out of curiosity.
Let me know what you think, and thanks. Herb
LikeLike
Herb,
I’m trying to think of an analogy – maybe a stone wall… all the stones line up at the Start of the wall, but that’s the only place, until you get to a door or window. Your DNA has fixed crossover points – we’re trying to “fit in” segments between two crossover points. Some may start at a crossover point, but many will start at random places, and then they all (mostly) end at a crossover (except those that span over more than one TG.
The issue at MyHeritage is that they use imputed SNPs to form the shared segments – that is they fill in more than was actually tested. This sometimes leads to longer shared segments than what we really have. It’s my understanding that they do *not* use the imputations when Triangulating – so the Triangulations are often shorter than the reported segment. And now that I think about that some more, we probably should shorten the shared segment when we find that. On the other hand, I just ignore a little fuzziness.
Instead of columns, I use rows to summarize the TG – I add a TG row (or bar) for each one. For the start location, I use .1 less than the first shared segment in the TG – say the first segment is 27.4 – 43.5; I’ll start the TG with 27.3. And then I go back to the previous TG (once I’ve assigned them to the correct sides) and, arbitrarily, have it end at 27.4 (there’s usually some fuzzy overhang….)
Don’t worry about small discrepancies – the data is fuzzy. Focus on the bulk of the TG as DNA from an Ancestor – treat TGs like the broadside of a barn.
Jim
LikeLike
Graham,
In my spreadsheet, Chr 15 doesn’t start until 18.3Mbp – the area is SNP poor and the companies do not show shared segments there. On my paternal TG 18.3 to 27.7 I have 75 segments from 4 companies; on my maternal TG 18.3 to 27.2 there are only 19… On my fathers spreadsheet with MH segments that I did for this blogpost, I have paternal TG 18.3-30.1 with 10 segments; and maternal TG 18.3-36.5 with 19 segments (all MY segments over 10cM)
LikeLike
Jim
I just realised what I put in. I meant to say DNA Painter has it marked from 20-25 and then 27-30. I don’t know why on earth I wrote 10-20
I am aware that we don’t get results until roughly 20cM on that chromosome.
Personally I have a only have a few matches between 20 and 30 and mine seem genuine although I will only be able to say that once I have proven the MRCA on them of course.
I’ll separately post about what I have been up to with my segments and matches. I went off at a bit of a tangent but it has been interesting.
LikeLike
Graham,
Re: your “seem genuine” comment. In general, all shared DNA segments over 15cM are true/valid/genuine/IdenticalByDescent(IBD). [with the possible exception of MyHeritage’s use of imputed SNPs which can result in longer false segments] To me, segments over 7cM which form Triangulated Groups, are genuine. Triangulation culls out almost all of the false segments. So whether the Match-segments are real (genuine), or not, is usually resolved with Triangulation. The second part of the puzzle that you allude to is finding the “correct” MRCA. And that is the real puzzle, and, as you point out, that is a genealogy issue – finding MRCAs with Matches. And not just any MRCA… I have a lot of Matches with Colonial Virginia Ancestry who have multiple MRCAs with me (only one can be right). The resolution is usually two-fold: crowdsourcing – what do most of the (widely separated) Matches say. There is only one correct Ancestral line, and, in general, most Matches will agree with the one line. The second method is Walking The Ancestor Back – either step-by-step from you to more and more distant Ancestors; or, given a particular MRCA, finding more distant cousins whose MRCAs are Ancestral to your given MRCA. Once you have a prospective MRCA, then you know that DNA must have come from one of the parents of the MRCA – and so on. So a combination of crowdsourcing an WTAB. Jim
LikeLike
Hi Jim,
Thanks for the blog, its an interesting variation on the Ancestry Clustering.
The only thing I would ask is the following:
When I am checking the triangulations for one segment on a chromosome I will often have triangulations for a subsequent chromosome. I assume you are numbering from 1 on each chromosome so we can’t use the same number but I think I will probably add a column for, let’s call them, Glocbal TG. ie. The group that covers everyone who triangulates with each other (the basis of clustering I guess). For this I won’t include siblings or children as they will link everyone together but it should speed up subsequent chromosomes because you will immediately know some people who are matches on a segment.
I am not sure I explained that very well !
This also looks like something that can be automated of course 🙂
LikeLike
Graham
Thanks for your feedback – a couple of things…
Although Clustering and Segment Triangulation both result in groups that are on one ancestral line, they are different. Clusters are based on grouping Shared Matches (with no information on segment location); and segment Triangulation is based on grouping overlapping DNA segments (with no information on the genealogy). Because our own DNA is fixed (recombination crossovers are fixed, and our Ancestors are fixed, and the segments our Ancestors pass down to us are fixed), I rely on segment Triangulation as a first choice, and use Clustering primarily at AncestryDNA (where segments are not identified). Segment Triangulation is much more precise, and each segment is taken into account – whereas each Match can only go into one Cluster, with gray boxes indicating otherwise. Also segment Triangulation at one Company, should result on the same TGs as Triangulation at any other company – Clustering can only be done within a Company, using Shared Matches. Clustering with different thresholds, usually results in different Clusters.
Segment Triangulation is actually done within the boundaries of the 45 or 46 Chromosomes. The TGs on one Chromosome are independent – physically separated from each other. In my process you can use the characters 1 to z in each Chromosome – in the end I establish a TG numbering system which includes the Chromosome number. For example: 06G2 is a TG on Chr 06, it starts at 60-70Mbp (as noted by the G) and it’s on my paternal side (as noted by the 2). This works for the expected 300-400 TGs. However, this is my scheme – anyone is welcome to use any system they want. Another way to identify 06G2 is: Chr 06 – 67.3 to 87.4Mbp – on the Paternal side.
Multiple Segments – when a Match shares more that one DNA segment with us, we must consider them independently. Most of the time the Common Ancestor for each segment will be the same – but not always. I have over 50 cases of Matches who share two segments – one from each of my parents. I have many Matches who share multiple Common Ancestors. I have a close cousin who shares 7 segments with me – 6 of them are from our close Ancestor, but one of them is from a distant Ancestor on another line. The outcome in one TG does not guarantee the outcome on a different Chromosome.
Segment Triangulation is a fixed mechanical process (no genealogy needed) – there is only one configuration – I hope it will be automated someday. Most programmers, however, don’t want to stop with just the groups – they want to identify which side they are on, and that process requires genealogy and judgment. Just automating the groups would save me 120 hours of tedious work. I would welcome that.
Hope this helps – if not, please let me know. Jim
LikeLike
I already informed Graham about the automation of the first grouping phase. The AutoSegment tool (https://www.geneticaffairs.com/features-autosegment.html) now has an additional excel sheet, inspired by this blog post, which automates the grouping. For MH, one then needs to manually verify each grouping, keep the group or break it into 2 groups. For GEDmatch, the groups are perfect since we already use triangulation data. For FTDNA, there is an option to employ ICW data to verify the groupings and that’s the best we can do for FTDNA since they don’t offer a triangulation feature like MyHeritage.
LikeLike
Thanks, EJ – I hope some “Triangulators” will try your AutoSegment tool, then manually verify each grouping, and leave comments here about how it went. Jim
LikeLiked by 1 person
HI Jim,
Thanks for the response. I think I didn’t explain well what I meant 🙂 … I understand the difference between Segmentation and the triangulation which you are doing …
Actually the main thing I was really suggesting was a speed up to your process. So while checking the triangulations of a particular user, I am making a note of the ones on other chromosomes. So when I come to that chromosome I already have some matches done.
Hopefully saves some time but maybe it would only give the impression of saving time ! 🙂
Cheers
Graham
LikeLike
Graham,
I’m not sure what “Segmentation” is. I started Triangulating DNA segments in 2012 (when FTDNA and 23andMe were the only atDNA tests offered); and I started this blog a few years later.
I thank you for exploring different ways to speed up my manual process. I, too, tried doing the process a Match at a time (I also tried working with only the Matches who only shared one segment). No matter which way, there was a lot of back and forth. I concluded (gut feel) it was best to just walk through each Chromosome. However, it is boring; and I often deviated from that “forced march”, and went down several rabbit holes. I tried to present something that was helpful, based on my experience of actually doing it. I welcome any and all improvements that folks find. And I encourage folks to deviate from my process and document a better one. Jim
LikeLike
HI Jim,
I meant clustering not segmentation. I mustn’t post in a rush, sorry !
I have been gathering the triangulations for a couple of hours today to try different methods but it wasn’t helped by the MyHeritage site running slowly in showing the triangulations today. And yes, you are right, it is boring 🙂 But it is still interesting to see the triangulation groups appearing.
I had a look at making the call direct to get the trianglations for a group of matches but didn’t have the time to give it a go.
Thanks again for the post, it has given me a lot to think about with MyHeritage.
Cheers
Graham
LikeLike
Graham – thanks for your positive attitude, and willingness to do the boring work. I think you’ll find the end result to be very beneficial.
Two things: sometimes I jumped to one of the shorter Chromosomes – which tended (for me) to go smoother; and sometimes I just skipped over an area that was being difficult, and came back to it later.
I hope you keep rough track of the time. Another total time will give readers another data point.
LikeLike
One thing I have just done to speed things up is to create a new field called URL which is made up of:
@concat(
“https://www.myheritage.com/dna/match/”,
the DNA Match ID,
?p=1&ps=10&sort=total_shared_segments_length_in_cm&siteId=xxxxxxx&individualId=xxxxxxx
)
You will need to grab the last part of that from a valid match view.
But you can then just ctrl-click or copy/paste into the browser and you will be straight to that persons matches without having to search for them.
LikeLike
Graham,
A good idea/process. I was trying to use a base Match and then “take care of” all the Matches with TGs from the base Match’s page. I was hoping I could drastically cut down on the number of Matches I’d need to look up. Theoretically, I’d only need to look at 350 base Matches… But it isn’t quite that easy to find one Match in each TG which will overlap and match all the other segments in the TG (particularly since at the outset, we don’t know what each TG looks like). Jim
LikeLike
Jim, in getting started with your Genome Triangulating, I wondered it you recommend including siblings? I also have a grandson on MH, but removed him from this process, because it seems to me that having him in the mix doesn’t really add any helpful info, but would add some other work to almost every chromosome. It seems to me that having the siblings, however, would be beneficial. I think the question in my mind was brought up by the Leeds clustering method, but in that case, identifying grandparents is the goal. Here, we’re to group segment matches and it seems probable that the sibling(s) matches will dominate the “long” segments, but I don’t see that as a problem.
One general question/comment. I had to look up the definition of “avuncular,” which appears to only apply to uncles, but I’m assuming in context as you are using it, you’re intending it to apply to uncles and aunts?
One clarification on my first comment above. When I mentioned the small matches I initially got when comparing my sisters, those were due to the setting I had on the chromosome browser. I’d been working on something else prior to looking at the sisters and had moved the browser’s “triangulated segments” setting to “2.” After moving it to 8 these ultra-small ones disappeared. Since they had all occurred at crossover points anyway, I had already assumed they were false.
I hope I’m not being a pest with my questions and comments. Thanks so much for sharing your experiences and your encouragement.
LikeLike
Doug, I do recommend using (not including) siblings. By that I mean I have a column in the spreadsheet for siblings and children. These relatives tend to share long segments with you, which are usually on the same side. So I note the groups that include a sibling or child. When I finish forming the groups, the next step is assigning them to a side, and aligning the segments on each side so they are adjacent. If you don’t have parents tested, the siblings and children can often indicate which side a group is on.
I meant avuncular in the sense of Aunt/Uncle/Niece/Nephew (my abbreviation is AUNN) – they, too, tend to have long, helpful, segments which span TGs.
Our conversations may also be helping others. Jim
LikeLike
Hi, Jim, I just found this recent blog (which I look forward to consuming) while accessing your site to share a MyHeritage experience with you and to see if you’ve tried this. I manage my two sisters’ DNA and just recently uploaded it to MH. Tonight while looking at a new match I just got a notice about, I decided to click on the trianglation symbol on the match list and see how the two of us matched with him. I don’t usually click on either sister when going through the triangulating pass here, so this was a kind of experiment. Well, we I only had 3 segments in common with this new person, and with my sister we triangulated on one of those because the sister didn’t share the other segments. Well, what happens if I through in the other sister? No triangulation! OK, what if I take the new match out of the picture? Wow, 48 triangulated segments, from 2.0 cM to 131.2 cM. There were a total of 6 < 7 cM segments and they all occurred at recombination points. I was curious if you've ever done this with siblings, or whether you have any interest in knowing more about this? One thing I can see right away is that this will be of benefit to me as I am trying to finish up my visualation process for the three of us by comparing with cousins on the paternal and maternal sides.
LikeLike
Doug,
Although you and your siblings each got 22 full autosomes from each parent, you and your siblings got different versions of each autosome. For any particular segment, you may be the only one to get it, you and one of your siblings may get it, you and both siblings may get it, one or both of your siblings may get it, or none of you three may get it. Some of my earliest blogposts go over how each of you get segments from your grandparents.
Yes – in my spreadsheet I have a column for siblings (I have 1), and children (I have 2 tested). They tend to share Matches (on the same side), and so they are good “tell-tales” – giving clues to adjacent TGs.
Since you have two siblings, you should use Visual Phasing (google it) – which maps the three siblings crossover points from their grandparents. It’s a difficult process, but then you have your grandparent crossover points mapped. And, YES!, you’ll see these same Crossover points between TGs. There should be about 37 of them (give or take a few) for each side (maternal and paternal). They separated your grandparents’ DNA segment in your DNA. Your DNA also includes about 37 *more* Crossover points from your Great grandparents; and 37 more from each generation going back. And your set of Crossover points is fairly unique, although some of them will be shared with one or both siblings. They are unique, and fixed (they don’t move or change in your DNA) – that’s why I like segment Triangulation – it identifies these Crossover points. They separate the TGs. Jim
LikeLike
Thanks for your response, Jim. Yes, I’m aware of the visualization process and have been through Blaine’s series a couple of times and have also tried working with the Fox visualization Excel tool. I have been working on these over the last few months. I’m pretty much completed with the identifying of the recombination / crossover points but still having some problems identifying the correct grandparent on some of the chromosomes. That’s what excited me about looking at the three of us with the MH chromosome browser and then adding in some of the cousins (one at a time) to help verify which grandparent is which on some of these segments. But, we’ll get there. I just want to say how much I’ve learned from and appreciate your writings. Thanks and keep at it.
LikeLike
Doug, Thanks for the kind comments. Segment Triangulation using a 10cM threshold should result in 300-400 Triangulated Groups – each one representing a segment of your DNA. Your 4 grandparents provided about 118 segments; your 1G grandparents about 192; 2G grandparents 266 and 3G grandparents 340 segments – so the 300-400 TGs would average out at about 3xG grandparents. Of course at that distance the DNA gets more and more random, and Matches with smaller shared segments within a TG will often go back more generations. Figuring out the crossover’s from your grandparents is a great start!
Jim
LikeLike
Your match list shows more than 14 thousand matches yet your numbering system of single digits and alphabet letters totals only 36 possibilities. I started with the first name on the list of matches (number one) and am just numbering them consecutively as I work my way down the list. I’ll end up with numbers in the thousands. I What am I missing?
LikeLike
Julian, I need to be more clear on two points:
1. Yes, the first Match-segment gets a 1; then, using that Match as a base, check for all the Match-segments which Triangulate with that base, and enter a 1 for them too – this way you form a group all of which have a 1. Any Match-segment which Triangulates with those, also gets a 1. The “1” group is a Triangulated Group (each of the Matches in that group should match many (but maybe not all) of the other Matches in group 1.
2. When you start with Chr 2, begin with 1 again. You should be able to identify all the the TGs in each Chromosome with a single character, 1-z.
In the end, the TG ID will be a combination of the Chr # and a letter, and each TG should then have a unique ID.
Hope this helps, Jim
LikeLike
Hello again, Jim,
You will be happy to know that my Chromosome 13 mystery with the 30 cM segment located within the 51 cM segment is solved — I think. I went back and took a really hard look at the 55 matches within my confusing Mr Arizona TG. Once again I found that these 55 matches were not matching the 46 people in the California Girl TG even though they were in the same segment range.
But, I noted a few new things. I had tentatively previously marked one of the Arizona group members as a possible early Wessling family member, a group which had its origins in NW Germany. About 3 of the Arizona group people had matches with a known member of my Wessling group (but not an Arizona group member). Now, it gets better. Some 23 matches of the Arizona group live in Germany or the Netherlands or Sweden, so this fact was telling. My mother’s people have been in the US since the 1600s and before that most of them were British. On the other hand, my dad had this one key group of ancestors from NW Germany.
I had absolutely discounted the possibility of this Arizona TG being on my dad’s side because so few (less than 10%) of my matches have been on that side. But, this seems to be a case where some of those NW Germans have passed their DNA down via Chromosome 13 to my Arizona group members. This solution also totally accounts for the fact that the Arizona people are not matching the California Girl people. They wouldn’t be expected to match because they are each getting Chromosome 13 data from different halves of the total chromosome. And, of course, it solves the problem of why didn’t any of those people who seemed to share the same segment were not matching.
Once again, thanks for helping me solve this problem.
LikeLike
James, The insight here is to follow the data! Triangulation is a fairly rigorous process that should result in a firm grouping. Conflicts are almost always caused by having the wrong Common Ancestor. When I started with Triangulation, I had a 2C with 7 shared segments, and I used the same CA (our common Great grandparents). But for one segment there was a clear conflict. When I really dug into the 2C’s Tree, I found we were also 5C on the other side. The CA on that side did fit in the TG.
On the positive side, as you determine the parent and grandparent for each TG, those TGs become pointers to the part of your Tree to look for more distant CAs…. This is why I refer to TGs and a Chromosome Map as valuable, powerful tools for your use. Jim
LikeLike
Hello Jim, and a Happy New Year! Thanks for all your good genealogy work. I have been giving your My Heritage strategy a go, starting with Chromosome 13. For me, this was a modest sized group with 142 matches there — and it is a group where I have a good amount of data on some of those matches. In doing this effort, I have already run across some questions that may also be common to others.
As a little background, I have 9,000 My Heritage matches. My father’s people came to the US from Germany in the 1850s while my mother’s people have been in the US since the 1600s, so I am getting well over 90% of my matches from my mother’s people. Further, I am generally familiar with my father’s matches — they have higher scores and are usually some degree of 2nd cousins. So, phasing is not usually a problem. My experience on Ancestry, FTDNA, 23 and Me, and GEDMATCH all tell me that my father’s people are only 5-6% of my total matches.
For my 142 matches in Chromosome 13, I have divided them into 7 well-clustered groups as we move through the segments from smallest to highest numbers. Four of these groups are quite small, with only 3-4 people each. My largest group has about 50 matches. One of these matches, California Girl, had a cM segment of 51 in this chromosome, so I used her as the key person in that group. She is a 3rd C 2R, descending from my GGF, so her score is much higher than one would expect. She and I have over 100 shared matches, and for C#13 I identified about 50 of them whose scores fall neatly in her same range, who all score over 10 cM on C#13. California Girl has a start at 27,399,818 and a stop at 73,065,967. I feel good about this group of matches.
However, within that same range there is another well-defined group with 50 matches. This group’s key man, Mr. Arizona, has a 30 cM segment going from 31,273,632 to 59,121,802, and there are about 50 people in that group who match with him. Yes, those numbers are right. California Girl’s score start earlier than Mr Arizona and end after his stop number. Yet, Mr Arizona and California Girl do not have a shared match with each other, and the matches within each of the two groups are also distinct from one another. It would have seemed that Mr Arizona and all of his matching people should have been shared matches for the California Girl people but it isn’t happening that way. Now, if it means anything, the very first lower scoring matches of the California Girl group, about 30 matches, are almost all shown to be shared matches with California Girl and are included in that group — but as the numbers of the start area get higher (but still within the range), more matches slide over to the Mr Arizona group side. I don’t think phasing is the problem, because I would have been aware of it if these people were on my father’s side — and further, none of them are showing shared matches with my higher scoring father-side people. So, my basic question is: “Why are there two distinctive groups within this same scoring range?”
I have another question which should be easier to answer. I have some good genealogy data on some of the California Girl matches. I know that she descends from my GGF. I also know that another member of that group descends from the father of my GGF. Does that mean that this entire group of matches is getting their DNA for that family branch as shown on C 13 from my GGGF — or perhaps an even earlier person. In other words, both California Girl and others in the group should be getting their C13 segment from the same source, however many generations back that may be.
Hope you can help.
LikeLike
James,
You work your way down a Chromosome, and group Match-segments as you go. You point out a lot match with California Girl. Select a new base Match near the end of that group and see who else TGs with this new base – often a TG is entended (made longer that way. This may or may not include Mr. Arizona. If California Girl’s matches match Mr. Arizona, then you have one TG; if not, you have two TGs. It is possible for the first segment in a TG to not Triangulate with the last segment in the TG – it’s OK.
Each TG represents DNA that came down to one of your parents to you. Alternatively that segment is from a parent and a grandparent and a GGGP, etc. So, yes, the ancestral line for each TG may extend 5 to 10 or more generations back. Somewhere, you “run out of” genealogy and cannot find another Common Ancestor going back…. Jim
LikeLike
Jim, Thank you for your quick response. I’m still a bit confused at the results I found. I would have thought that when one of my matches had a 31 cM segment on a certain chromosome within the segment range of another match who had a larger 51 cM segment that the 31 point segment should be identical and that the two of them should match — but they don’t. And it happens that way sometimes.
One thing I have learned is that to establish a solid TG you not only need to identify a common segment area on a specific chromosome but that the matches within the TG need to be shared matches. That is, you can have two matches who share a given segment but those two people will not necessarily be shared matches and thus are likely to be in different TGs.
LikeLike
James,
You’ve hit on the crux of the issue with Triangulation – landing on the right chromosome. A 31cM segment totally within a 51cM segment, must either Triangulate with the 51cM segment or be in a TG on the other chromosome from the 51cM segment – the 31cM segment is too large to be false. Look for other, smaller segments that TG with 51cM segment or the 31cM segment – they may for two different TGs – on opposite sides (one paternal, one maternal). In a few cases, I’ve seen the smaller segments “stitch together” the two larger segments. This means the one or both of the larger segments isn’t really the full segment it appears to be – it’s been made larger by imputed SNPs which are not considered in TGs. … Jim
LikeLike
This has inspired me to have a go at creating TGs from my own data. I have a DNAGedcom download of triangulations at MyHeritage, which makes it easy to automate the process. To acquire the download required running the DNAGedcom client for several weeks last summer, so it is not necessarily quicker than your method but a lot less work. The basic algorithm is:
1. Select a segment
2. Find all the segments it triangulates with
3. Find all the segments they triangulate with
4. Repeat (3) until no more segments can be added and the TG is complete
5. Repeat (1)-(4) with another segment until no segments with triangulations remain.
It took me a few hours to code yesterday afternoon, but I now have 203 TGs based on segments over 10 cM, or 375 based on all segments. There are numerous places with more than two TGs, which I have yet to investigate properly, but this accords with the little bit of manual analysis I have done which found more than two TGs in some places using MyHeritage’s triangulation. Now to work out which ancestors they are associated with…
LikeLike
Actually my process (or algorithm) is a little more efficient in that it eliminated step 3 – there is no need to determine all the pair-wise Triangulations. Once a TG is formed with two segments (plus your own), all you have to do is add segments which Triangulate with one other segment (in addition to your own). I often do this twice, just as a Quality-Control check, and to avoid errors. But not for each pair – which, in fact, won’t work in many TGs: the segments on one end (of the TG) may not overlap the segments on the other end enough for a match.
Based on the 7cM threshold I used over the past 8 years, I have 372 TGs – so it appears from that metric, you are definitely on the right track.
One word of caution. It is important to Triangulate *segments*. Many of the processes I’ve seen Triangulate *Matches*. Triangulating Matches works when those Matches share only one segment with you and the other Matches. When they share multiple segments, a faulty “Triangulation” can occur (not based on overlapping segments). I’ve used DNAGedcom for years – it is a great program for downloading data and for Clustering (which is not the same as Triangulation). Triangulation of shared DNA segments treats each segment separately. This is important for endogamy. Clustering does not handle endogamy well, because it does not handle multiple segments well.
I’d be interested in your observations of multiple TGs. My experience is that these occur because of close relatives (parents, children, siblings, avuncular, 1C and 2C) who tend to share DNA larger segments that span TGs. In an automated process, this can be fixed by making one pass without those large segments to get the TGs set; and then making another pass to add them back in (these long segments really need to be indicated in each of the multiple TGs they Triangulate – same problem with multiple segments in Clustering). These close relatives/large segments are very valuable in assigning “sides”, so they need to eventually be included. All of this is why, so far, I prefer manual Triangulation. But any process that automates the bulk of Triangulation would sure avoid a lot of the tedium of my process. I did a comparison of using both processes (Triangulation and Clustering) on all of my FTDNA Matches/segments>7cM. The results were VERY close. When I manually reviewed the multiple-segments issues, I got very close to 100% alignment. Meaning that both processes point to a unique Common Ancestor – a very important and valuable outcome. I used Clustering extensively at AncestryDNA, because they did not provide segment data; but for the other companies (with segment data), I prefer Triangulation because it is more precise and uses all the DNA data (and it provides a map of my Genome which lets me add new Match-segments easily). For me, the end game is finding the most distant Ancestor I can for each segment (TG) of my DNA.
LikeLike
Determining all the pairwise triangulations is no work, it’s what the download gives me. Step 3 is necessary or you only have the triangulations with one segment, and may only get one end of the TG as you describe. You need to keep expanding the TG by finding every segment that triangulates with any segment already in the TG, and repeating that until there are no more to find.
My multiple TGs can’t be due to close relatives as my top match is a 4C1R at 82 cM. I suspect it is due to the issue you mention in your post of imputed SNPs being used for matching but not triangulation.
LikeLike
My point is that my process addresses every segment – each one pairs with others, or it is false (actually a segment over 15cM may be the only segment for that particular area of a Chromosome). If every segment is analyzed, that’s all you need. Once a segment is grouped in a TG, it does not need to be revisited. A segment at the other end of a TG may well match previously triangulated segments, but those previously triangulated segments don’t have to be used as a base segment. But this discussion is mute, if all the Triangulations are automated. It just doubles the work (A to B *and* B to A), but what’s a few minutes to a program.
LikeLike
I thought my process was clear on this, but maybe not. Pick a segment as the base; annotate all the segments that Triangulate with it; then pick the next un-annotated segment as the new base; and repeat. In this way you “touch” every segment – each segment is either a base segment or a segment which Triangulates with a base segment – all are Triangulated, except a few that don’t Triangulate with any others. If these overlap a TG on either side but don’t Triangulate, they are probably false; if they stand alone, they may be the only segment in a given space on a Chromosome = wait for more info.
LikeLike
Hi Jim,
Excellent article as always.
Are you keeping a separate spreadsheet for each testing company?
How will you deal with new matches as they accumulate at each company?
LikeLike
I have only one set of DNA – the data has to conform to me, no matter which company it comes from. My crossover points, and thus the TG segments are fixed. So I put all the segments in one spreadsheet.
As I get new segments, I tag them with today’s date, add them in, anywhere, and then re-sort the whole spreadsheet by Chr + Start. I then just search the spreadsheet for today’s date and adjudicate each one – does it Triangulate with a Match on one side or the other. Just to be sure, I usually triangulated with two Matches.
The hard part is determining which are the new Matches. It’s easy at GEDmatch and FTDNA. For the others you could ask Genetic affairs to tell you the new ones. Another way (fairly complex) is to compare your current spreadsheet with a new down load and use the ones that don’t match up. Jim
LikeLike
This is a different spreadsheet than the one you used for 23 and Me. I was about to begin working on triangulating and grouping my 23 and Me results. For someone like me who is just getting into segmentology (I’m reading all your posts which I find to be excellent) would it be best to use this method and spreadsheet for any triangulation grouping of tests from the different companies I’ve tested at? Thank you so much much for all your posts. You are a true asset to the genetic genealogy community.
LikeLike
Bob, Thanks for the kind feedback. Any spreadsheet will do – Just download your segments at 23andMe and use that spreadsheet. Add columns when you need them. Review the blogpost and the figures to see the ones I recommend. The order doesn’t make any difference – in fact, I sometimes move some the columns around. For grouping, the one you’ll use the most is one for the ID# – that column is the key. I’d do one company at a time…
LikeLike
Yikes! Good thing that I am pretty much a hermit due to covid :-)!
LikeLike
I’m in category 1b, and we’ve only rarely left the house. This project took my mind off the boredom.
LikeLike
I’m 1b, too. I do leave the house four times a day … to walk the dogs. If I walk them a mile, they will take a nap and I can get back to genealogy until time for their next walk! I’m definitely a spreadsheet person, so this is right up my alley. I just finished grouping the segments on Chr 20. Guess I’ll try a little bigger chr next. Your instructions are so specific and easy to follow. Thank you for all you do for the genetic genealogy community and for sharing with us.
LikeLiked by 1 person
Jim,
I love your postings and have found them extremely useful. Especially your thoughts on the smaller segments. I admit I haven’t thoroughly read this posting on your process, but are the results much different than what you would get using Genome Mate Pro?
I’ve used GMP for a number of years and love how it organizes all my data. However, I rarely find it recommended. Sorry if I’ve stumbled onto some turf battle or something. That’s why I chose to email you rather than make a comment on your blog.
Thanks for your willingness to share so much of your thinking and process.
Jennifer Clark Sent from my iPad
>
LikeLike
Jennifer, Thanks for your feedback. I recommend all the tools – whatever works. Each person has their own favorites. I was several years into my Excel spreadsheet when GMP came out. I tried it, and it was good. But the thing is, I didn’t want to abandon my spreadsheet and start on a different platform. I was the same way with Clustering – all three programs work, and I just settled into one of them for most of my Clustering work. With all the data that is still pouring in, it’s difficult to keep up. I’ve stopped tracking most segments under 10cM. There is good information in some of the small segments, but I’m pretty much done with gathering and analyzing segments – my segment map is done, and my focus now is on digging out Common Ancestors.
With GMP have you determined 300-400 TGs – all adjacent to each other, and covering all of your 46 Chromosomes? If so, then GMP should be recommended more. If not, then I would recommend my process in the blog post. Within a finite amount of work time, you can determine how virtually all of your DNA breaks down into segments from Ancestors. What’s left is determining the correct Ancestor for each TG segment.
LikeLike
Jim, I work primarily with my mother’s DNA test. Using GMP I have been able to map her paternal side with about that equivalent number of TGs. Her maternal grandparents were recent immigrants from an area of Germany where few people have DNA tested or where few people migrated to other continents – so finding any match is pretty difficult.
What I like about GMP is one database includes tests for my brother, a maternal cousin, and myself, and that helps me to at least be able to determine which side a TG belongs to.
I started with GMP very early on in my DNA work, so, like you, I’ve stuck with what I’m familiar with.
I do have a number of TGs that I haven’t found an ancestor for – but that’s because the groups all point to colonial New England. Either I can’t work a match’s tree back that far, or there are several possible shared ancestors.
LikeLike
Jennifer, I cam empathize – my maternal grandmother’s father was an immigrant from Scotland (CAMPBELL) and her mother was an immigrant from Germany (WEHRLE). I have only a handfull of cousins from this lines, and a number of TGs with no known Common Ancestors, and aa few TGS which are blank (no matches with that segment).
I have spreadsheets for my brother, uncle and father – sometimes I’ll combine then all into one and compare information. But I always separate them apart again – the segments are not compatible! As I pointed out in the blogpost, I have columns to indicate when one of my Matches also match a child, sibling, parent or close cousin. These close relatives tend to share large segments of DNA with me which may include several TGs. They often help in linking TGs end-to-end.
In the end, it’s the quest for Common Ancestors with Matches that is the key (mapping the segments is done). I’ve long since documented all of the obvious CAs. I’m now using the Groups to help me find more – often I need to extend a Match’s Tree to find the CA.
LikeLike
Jennifer, You may already be there, but MyHeritage has more European DNA testers – I have 124 matches in Germany and 84 in Switzerland. I wish there were more.
LikeLike
Yes, I do have my tests at MyHeritage. And that site has been somewhat useful for my Pommern ancestors. A lot of folks from there also migrated so I can find their descendants at other sites. Fortunately, I’ve been able to document my Wuerttemberg ancestors through church records, many lines to the late 1500s. They are the group with few DNA matches.
Back to MyHeritage – their marketing must be paying off – most of my good matches for all branches of my tree over the last 6 months have been there.
LikeLike
Hi Jim,
Extremely nice roadmap – just WOW. I’m still working on clusters and collecting segment data for known ancestor matches using Excel. Haven’t started inputting DNAPainter.
120 hours? And on the 6th day you rested?
LikeLiked by 1 person
Randy, On the about the 50th day… I had a libation and started collecting my notes to write the blogpost;>j I’m a big fan of Clusters – particularly for Ancestry data where there are no segments. But I’m convinced the “end game” has to be linking Ancestors to Segments – it’s the only way to smoke out the NPEs. Also, to some extent, TGs really help with endogamy, because each segment (TG) stands on its own merit – it comes from one path down to me. It’s still a hard puzzle to solve, but a lot of Matches in a TG all on the same path (somewhere), sure helps.
About a year ago, I inputted my 372 TGs into DNA Painter. It gave me a nice picture of what the DNA said, but it’s just that – a different picture of the same thing. The chromosome painting is the same data as the TG segments – just a different way to view it. For me, it’s easier to work with (sort, compare, manipulate) the spreadsheet. Others get a lot more out of the visual picture.
LikeLike
Jim, is there a way to take your triangulation groups from the spreadsheet and input them into DNA Painter, or do you have to go to each site, find each match, and input the data from the site if you don’t want to key in the match data from the spreadsheet?
LikeLike
Richard, Yes! I’ve done it. Go to the Tools page and read about Cluster Auto Painter… Jim
LikeLike
Jim, there seems to be a step or two missing here. The Cluster Auto Painter page talks about choosing a clusters HTML file, with a choice of sites and to generate the HTML file, none of which seem to be “I have this lovely spreadsheet sorted by triangulation matches that I want to paint.”
LikeLike
Richard, Sorry – it’s been a while. To import from a spreadsheet, use these instructions: https://dnapainter.com/help/import – and a tip of the hat to Jonny Perl who just reminded me of where this was.
LikeLike
Wow! Thank you – guess I know what I’ll be doing for the next few weeks
LikeLike
Watchinit, Settle in, and good luck! 120 hours is about a month at 4 hours per day; or 8.5 weeks at 2 hrs per day. I was into gardening this past summer, so my pace was only 2-3 hrs per day…
LikeLike
No genealogy for me from Mar-Nov – demands of the hobby farm, but it’s good. Maybe by the time I get back to to the mega match project, someone will have written software to automate the process. Happy New Year! – hope 2021 is your best year yet.
LikeLike
Hi Jim, very interesting blog post. I am not a segment expert but it seems this process is more or less an offline DNA painter profile using Excel? Some thoughts. Are you correcting the basepair positions for FTDNA segments, since they are based on build 36? I have shared segments with the same match on FTDNA and MyHeritage and some segments are not overlapping. Also, it seems like a large portion of this work can be automated? As you perhaps know, I’ve tried that as well and came up with AutoSegment: https://www.geneticaffairs.com/features-autosegment.html. Doesn’t make the comprehensive spreadsheet, but instead create segment clusters, probably similar to your TGs. It’s also possible to combine the four sites that provide segment data, and to make data more comparable I perform a liftover for FTDNA.
I am currently finishing up a version for FTDNA that is using ICW data to verify overlapping segments. Segments that overlap, but whose matches are not shared matches, are not valid and won’t make it in the same segment cluster.
LikeLiked by 1 person
EJ – Thanks for your comments. I am a long time fan of your work. As you can tell from my post, my focus is on the whole genome. I’ve been a genealogist for over 45 years, and have gone about as far as a can with paper research. When atDNA came along in 2010, I used it to find Common Ancestors for a few years, and then got hooked on segments and the bigger picture. Mindful of brick walls and NPEs, I am focused on linking my DNA segments to my deepest Ancestors. I want the full map, and I want to build it up a generation at a time.
This is an offline process, with Excel, I’ve been developing and using since 2012. I have not corrected FTDNA segments. I have all my FTDNA, 23andMe, GEDmatch and (most of) MyHeritage shared segments in a one spreadsheet (over 20 thousand rows). My 372 Triangulated Groups are heal-and-toe from beginning to end of each Chromosome, with only 10 or so, small gaps. The ends of these TGs are often a little ragged – FTDNA using different build; MyHeritage inputations; etc. But my thesis is that my crossover points are fixed, and almost all shared segments will fit within them – with fuzzy ends. I’ve mapped my DNA segments – I now want to verify where they came from (and thus verify the genealogy). I think TGs and Clustering are great tools for this objective.
Some have tried to automate this process. GEDmatch did a pretty good job; and MyHeritage and 23andMe have very accurate TG indicators – unfortunately there is still the issue of mapping all this data. As with Clusters, multiple shared segments must be dealt with. And there will always be the issue of a Match who shares multiple Common Ancestors with me. It always comes back to which Ancestors go with which segments.
I tried to help FTDNA several years ago – they tried, but never got the brass ring. I believe overlapping FTDNA Segments PLUS being on the ICW list with each other makes a solid TG. I Clustered ALL of my FTDNA Matches (7cM threshold), and found an almost 1 to 1 correlation with my 372 TGs (after I analyzed the the multiple segments independently and made some adjustments in Clusters) This convinced me that both TGs and Clusters are focused on a Common Ancestor.
I think one of the important outcomes of Clustering (and TGs) is the identification/elimination of false segments.
LikeLiked by 1 person
Hi Jim, thanks for the elaborate reply. Glad to read that you also found that ICW data can serve as evidence for overlapping. I think that’s the best you can do with FTDNA, similar to their matrix system. I still think there is some added value supplying the corrected FTDNA values since for some chromosomes a 10 cM segment for FTDNA wasn’t overlapping with the same 10 cM segment on MyHeritage. That will be impossible to place underneath each other unless you correct for it. I think my AutoSegment tool would be an excellent candidate for your Excel file. All the necessary data is there, (corrected) segment data, overlap, etc etc. The only thing different is the overlap calculation, I don’t use Mbps but instead, calculate the overlap in cM using genetic maps. Doesn’t really matter too much, but in some cases, there is quite some difference. In that case, the TGs are probably not separate but should be combined to reflect the underlying recombination characteristics. Let me finish my new tool in the upcoming days, I’ll spend some time seeing if I can replicate your Excel file automatically.
LikeLiked by 1 person