I presented “More Segmentology” today at the East Coast Genetic Genealogy Conference. I was questioned on a slide grouping segments into a Triangulated Group, and it appears there is a debate about this. I’d like to have your input on this.
Here is my slide:
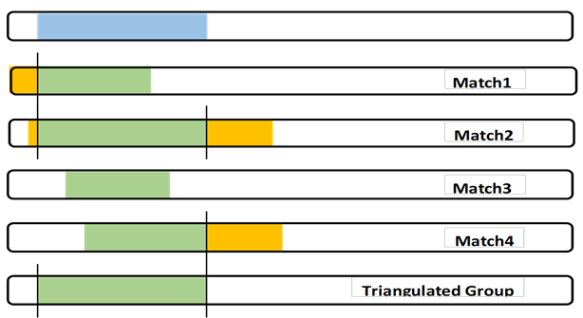
I show 4 Matches with overlapping Shared Matches with me on one side (parent). This is the definition of a Triangulated Group, which I showed in the bottom Chromosome – in green. What we can “see” is only the Shared Segments from Matches 1 to 4 in green. I contend that Matches will rarely have segments that are exactly the same as my segment. So for the purpose of illustration, I guessed that their segments from our Common Ancestor was almost always different – that sometimes their segments started to the left of mine, sometimes to the right of mine; and sometimes the same ending, and other possibilities shown. In fact, I have tested this at GEDmatch where I could Triangulate with each Match as the base, and sure enough, they had their own, different, Triangulated segment. I went on to claim that my segment (from our Common Ancestor) started where the Shared Segments had their earliest start; and ended where the Shared Segments had their latest end – as shown in the green Triangulated Group segment above. The start and end of the TG defined my segment. Some others contended that the Triangulated Group segment should be shown as only the green that was common to all 4 Matches – like the space between the two vertical blue lines.
I don’t know of any Scientific Paper that defines the boundaries of a Triangulated segment. So I am interested in your perspective, and why.
[08Aa] Segment-ology: Boundaries of a Triangulated Segment by Jim Bartlett 20250914

Jim, Lots of great points made here, I just want to add 2 points. 1. DNA can come from a couple, just his side or just her side which both go back to a couple a gen. further back. By not including Ancestor names or match relationship, you are missing out on this layer of analysis in your graphic. 2. This discussion points to the depth and breathe of a DNA connection by the match info we compare. I think we may need to define that with common terminology/labels with exact analysis can’t be done. Ancestor Line connection vs. Shared DNA Connection vs. Shared DNA segment . This can be discussed at length and some in our field never use DNA segment info in their analysis. That has always puzzled me with the GPS standards!! Someday I think your type of DNA critical analysis will be best practice, but when DNA matches can’t provide full segment info, then our analysis is our best guess with the matches we compare as of that date in time.
I compare all this the paper records when you have a marriage date, place and then find an official record that verifies it all. It was all probable until you could prove it with a “primary record”. You are always making us stretch are skills and I thank you!!
LikeLike
Cary, There is often confusion about what an MRCA is with resepect to how we get DNA. Some people prefer the specific Ancestor person who passes down the new segment; I prefer the Ancestral couple. I prefer the MRCA couple because that is all we know when make the genealogy connection (avoiding the cases with different spouses). In other words I descend from a child of the couple and my Matches descend from other children of the couple. Without more info, I don’t know whether the DNA segment came from the father or the mother of the children; and in most cases it may be a long time before I can ever figure it out. And whenever we determine an MRCA couple there are two options: 1. the TG segment (= to my DNA segment) was first formed in one person in the MRCA (initially as an imbedded segment in a whole chromosome, and it had to have a crossover in it – between the parents of the man or woman in the MRCA couple); or 2. the full TG segment is actually also in a parent of the man or woman in the MRCA. This then gets us into the concept of the Most Distant Common Ancestor (MDCA) couple who had the full TG that I got. My graphic was intended to show MY DNA segment (= the TG segment). That segment had to exist in my parent, in my grandparent, in my great grandparent, etc. all the way back to the man or woman in the MDCA (and no further back). When we go back another generation from the MDCA, the segment is then two segments, each one from a different grandparent of the man or woman in the MDCA couple. The result of this “traveling TG segment” is that my Matches can be anywhere on the path to the MDCA – 1C, 2C, 3C, etc.
I focused on segment Triangulation for years, thinking that was the hard part and a full chromosome map of my 372 TGs would be the key to unlocking everything… WRONG. As it turns out it’s very difficult to find many MRCAs at the companies that have browsers. On the other hand, at Ancestry I have solid paper trail links to every one of my known Ancestors out to 5xG grandparents (6C level with good ThruLines to them all). But I only know the TGs for some of them (who have tested elsewhere or uploaded to GEDmatch. It’s frustrating.
Incidentally, we can find full segment info with Matches at GEDmatch, if we create TGs for each of them…
Jim
LikeLike
you are so right Jim. It would be far more helpful if Ancestry would provide a chromosome browser in its latest upgrade instead of all the guff about ethnic origins which tends to change every year or so.
LikeLiked by 1 person
I’m with you
LikeLike
Thank you, Jim, and thank you to all the commenters for your insights, because I have always found this confusing. It is actually the reason why I stopped trying to work with triangulation groups a couple of years back. I might have 10 matches with large overlaps, but only a tiny area that they all absolutely overlap each other. The idea of “fuzzy” segments was not helpful. How fuzzy is fuzzy? The comments you’ve all made make me think that maybe I’ll take another look at this sometime. I like the suggestion that maybe we need different terminology for the two approaches. They are two different ways of looking at the data; both approaches can both be useful. Using different terms for each one could help keep our thinking and communications clear.
LikeLike
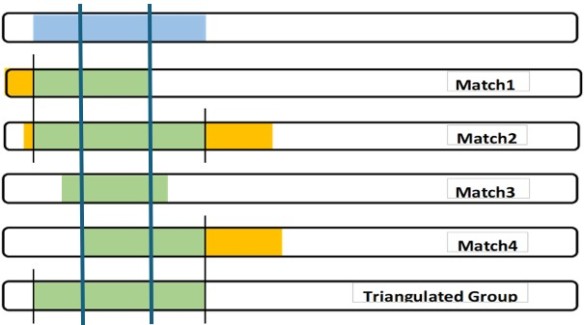
It depends on your purpose in identifying triangulated groups. Triangulated groups are groupings of YOUR matches, meant to point to an ancestral couple common to YOU and the other matches in the group. True, your blue segment came to you from a Most Recent Common Ancestral (MRCA) couple, and some of your matches have smaller pieces of it. All of four of the matches in your illustration (Matches 1, 2, 3, and 4), have a piece of it and are descended from that same MRCA. But the MRCA ancestor responsible for that blue segment got it from his/her parents, who got parts of it from their parents, etc. So the blue segment is a composite of smaller, older segments from earlier ancestors who have descendants that are related to you through other paths. To illustrate, let’s divide your blue segment into three parts, A, B, and C. Part B is the portion between the heavy blue lines that the more restrictive definition would say is the triangulated segment. Parts A and C are the portions to the left of B and to the right of it, respectively. Part A may have come to you and Match 1 and Match 2 only because you are more distantly related to them than you are to Matches 3 and 4. You may want to consider your matches who share Part A to be a different Triangulated Group, particularly if, on other chromosomes, you share segments with them and not with Matches 3 and 4.
So it’s a matter of granularity. If you’re just starting out, you may want to define the triangulated segment as the one that’s equal to your blue segment. But if you’re working further back in time, the more restrictive definition becomes more valuable. Defining two separate TGs in the above illustration may be helpful in finding the bio parentage of a gr-gr-grandfather, for example. I think this is parallel to Walking Back the Clusters.
Bill Josey
LikeLike
Bill, Thanks for your input. This is a two part issue – segments and MRCA. The TG segment is the best I can do with the given data – and clearly we all got DNA from a Common Ancestor. I agree that all segments can be subdivided and that goes, partly, back to how far back can I do the genealogy. I sometimes struggle to find a CA for some TGs (Matches with segments tend to be from companies with out much genealogy help).
I agree with granularity and try to walk the Ancestors back on each TG segment – with varying degrees of success. As you say – like we do with Clusters.
This might be a good topic to discuss on our next Adv DNA SIG Zoom….
Thanks again, Jim
LikeLike
For me, it’s the longer representation in your first slide which is of main interest. The smaller segments shown in the second slide represent different “windows” on the piece of DNA that is of interest.
That very smallest segment in the second slide just shows the single, isolated segment of DNA which everyone in the group shares. That may be interesting for some reason, but as far as researching the ancestral history of the larger expanded segment in the first slide, that’s the “group” I want to spend my time one.
LikeLike
Glen, Nicely stated – thanks – I agree with you. Jim
LikeLike
Hello Jim – hope the conference went well, I’ve registered but haven’t had a chance to watch the recordings yet. Looking forward to it!This is a very interesting question. When I first start working with people to get them to understand how TG’s work I would generally tell them to focus on the part of the segment that everyone shared. This was to ensure they look for the same common ancestor for all the group.I totally agree that the start and end positions would rarely be the same as your segment. They are often extended on each side due to differing recombination points for any of the matches.
From the testers point of view the entire length of the segment is of interest and this is how I define the boundaries of each of my identified triangulated group’s. In your example, Match 2 shares all the same as your blue segment so I would use these extended locations to analyse the other matches in that broad location.
I often find that when analysing a group of matches that have an overlapping area like in this example its best to work on the largest match to look at triangulations along the segment. You will often find several sub segment TG’s along the length of it. The largest match like Match 2 could also suggest a closer relationship. In the second image you could look at it as 3 sub groups. However, in the example all 4 triangulate in the centre which indicates a shared MRCA so the extensions to the left and right must also come from that same ancestor (if they were 2 ancestors then there should be a split). This affirms it is appropriate from the testers point of view that the broader TG area is defined by the start and end of the blue segment.
However at times a match like Match 2 may be a close cousin and the others more distant relationships. In that case there might be several TG’s groups along the segment, but there would usually be a gap (or very close start and ends) which is not the case here, where all four matches did not match each other. In those cases the 2C might be sharing your great grandparents, whereas the groups on either side likely only one of them.You would expect as more matches are added to this group of 5, sharing even more distant ancestors, that this pattern would start to emerge here as well. In the strictest sense of what we are defining, all 5 testers are all members of the same Triangulated Group. However, the ‘shared segment area’ of the ‘group’ is limited to the smaller location area that all 5 share.The boundaries for that specific TG from the ‘individual tester’ point of view would be defined by the extended start and end locations. Each tester in the group would have different boundaries as they would match each other differently.Perhaps we do need some better definitions. I know the concept is confusing for many, particularly when looking at TG’s on MH where imputation can create problems in analysis.Looking forward to hearing others thoughts.Veronica
LikeLike
Veronica, Thanks for your feedback – I’m with you all the way. Most of my 372 TGs are pretty cut and dried; but some are weird. There are some with vestiges of a split – some on one side, some on the other side, and many spanning most of the TG segment. I think with my 372 TGs, I’m backed up to about the 8C level – it’s going to be very hard to find records further back. I keep reminding myself that all of our segments are probably subdivided going back (clear back to my very small Neanderthal segments…).
Jim
LikeLike
Hi Jim,
A bit more on my POV.
If I look at the segments, I maintain that the triangulated segment for this group is represented by the bottom panel. It is the only segment shared by all 5 testers. The boundaries of that segment will change as we add or subtract testers that have an overlap.
Now if we consider the concept of triangulated group, we may have a different interpretation. Here the focus is on group – the collection of testers that have significant overlap in their segments. This is where the top panel becomes the answer. It is the group of testers that, based on overlapping segments, likely inherited those segments from a common ancestor. The question now becomes how much overlap is enough overlap to gain admission to the group. We are not likely to admit someone who had only a 2 cM overlap with any of the testers. Perhaps since our testing window is 7 cM, we should demand a 7 cM overlap to admit a new member to the group. Or perhaps 14 cM considering the window is 7 cM for each tester.
I do agree that we usually are interested in making conclusions about the triangulated group (testers) since that is what is of genealogical interest. Therefore the top panel is what we most often see. However when we speak of triangulated segments, then perhaps we should use the concept of the bottom panel.
JM
LikeLike
Thanks, John – We agree the Group of Matches should all be looking for the same Common Ancestor – hopefully we can help each other toward that goal.
I will also use the first panel, full segment, result populate my Chromosome Map; and contribute SNPs to any group effort to build the DNA of an Ancestor.
And I need to be careful to not try to convince my Matches that they, too, actually have this TG…. The TG segment is mine, and my Matches should have their own TG segment, with an appropriate overlap with my TG. Jim
LikeLiked by 1 person
Hi Jim,
Yes of course we are working in the same direction!
This is nothing I want to be dogmatic about or press further. I do think it was something worth discussing. Thank you for graciously using your forum to enable a conversation that’s not possible at a conference and opening it up to more people.
JM
LikeLiked by 1 person
John, I like your POVs – and I agree it depends on the concepts involved. I got into segment Triangulation to group all my Matches around my segments. So my concept is the TG represents (let’s me “see”) my DNA segment. And, borrowing from the genetic genealogy foundation that shared IBD segments means a Common Ancestor, this results in the TG Matches will have a CA. So, for me, a TG groups segments to define my segment (including crossover points); and the TG Matches will share a CA with me => takes me back to my genealogy roots.
I need to refer to *my* Triangulated Group segment more; it is different from the TG each of the Matches would have. The Matches are all in the TG, but don’t share the exact same TG segment.
Thanks for your feedback.
Jim
LikeLiked by 1 person
Jim, I can see why people might quibble with calling the fullest extent of the triangulations among matching pairs TG. But I think it’s helpful to think of it as the ancestral segment. In my published works I adjust start and end points owned by one member based on what is shared with the others in the TG. I’ve not taken it a step further to identify the entire expanse as a TG. I think it is more helpful to think of it as an ancestral segment (and give the visual) than to only show matching pairs numerically.
LikeLiked by 1 person
Patti, Thanks for your input and experience I got to see your experience up close when we taught together at IGHR…
I’m beginning to understand the perspectives… I look at the full blue TG as my segment of DNA, which had to exist in every one of my Ancestors back to the Common Ancestor we all share. Each Match in the TG got a different, but overlapping segment from the same Ancestor – they would each form a different TG.
For me it’s essential to use a spreadsheet to collect all the shared segments, sort them and then Triangulate the overlapping ones (tossing the smaller IBS ones that don’t Triangulate). I then resort my spreadsheet into two sides and then sort each side by chromosome and start to clearly show the TG groups of Matches – from there it’s a tool to work with the Matches in a TG to determine our CA.
In the end it’s fun to paint the segments, but I usually get stuck in the spreadsheet trying to find a consensus of Common Ancestors. My segment spreadsheet has over 25,000 rows of segments, but they condence down to 372 TGs (plus some IBS segments).
Jim
LikeLiked by 1 person
You question seems to be whether to use the intersection of union of matching segments in your Triangulated Group. The answer is likely Intersection but can depend on what definition you are giving to the triangulated group segment.
Firstly, it seems you need 10 segments there for the 5 test kits being “matched” or triangulated. For a triangulated group, it is important that all 1:1 pair match-ups be represented with their overlapping matched segments. If you do not have 10 overlapping segments, it is not a triangulated group. You may have a different definition for this term in your blog; I cannot recall. I follow Blaine’s 2016 definition (see ISOGG wiki triangulation page).
Second, how close (in time/generations) are the MRCA(P) for each of these 10 pairs? As an extreme, if one of the matches is your sibling, then the overlapping segment would likely be very large between you two. But how much of that segment is due to the MRCA between all matches and just the closer in time MRCA between you two? So unless each match is from a different child of the same MRCA(P), then using the union potentially brings in portions of a segment not attributable to that common MRCA(P). Practically, if all the pair wise MRCA(P)s are distant and close to the same generational distance, then maybe this is not so large an effect.
Finally, what is your use or definition for this segment? Are you trying to associate a portion of your DNA attributed to a particular ancestor? Or trying to define a portion of each matches DNA to a particular ancestor? Maybe a Union is more appropriate for the former and intersection for the latter.
All the standard caveats of no endogamy, having strong evidence of knowing your complete genealogy, etc apply.
So, in general, the intersection using all 10 segments is the way to define the area on everyone’s DNA of a segment attributable to a specific MRCA(P). Independent of how close the MRCA between each pairing of the 5 matches there is. But practically, if you all have a same / similar distant MRCA(P) between all matches, then the union of overlapping segments of just matches to you could be useful in your particular DNA “painting” trying to identify the source of your particular DNA.
LikeLiked by 1 person
Randy, Thanks for your feedback. Yes, it does depend…. I should note that my definition of a Triangulated Group is two fold: 1. A Group of Matches who all should share the same Common Ancestor; 2. A segment I got from that Common Ancestor, which defines two of my crossovers; can be placed in a Chromosome Map and forms an atDNA haplotype in my Common Ancestor .
Yes, all of the segments are Triangulated. Actually, once you form a Triangulated Group with two separated cousins, you can add new Match segments to the TG by only comparing to one segment already in the TG (I usually confirm two to be sure) – I’ve checked this and the other with sufficient overlap always also Triangulate.
At this point I don’t really know the Ancestors along the TG segment. They are usually found later, by working with the Matches in the TG.
My use for a TG is to map all of my own segments (the full blue one in the post); and then to link it to an ancestral line via 2C and 3C and 4C and 5C etc. as far back as I can – trying to determine the source.
Thanks again, for your input. Jim
LikeLike
Thanks Jim for the follow-up!
LikeLiked by 1 person
Ciao jim ti mando una foto della mia triangolazione puoi dirmi che significano quei numeri?
LikeLike
Hi Jim,
I enjoyed your presentation at the ECGGC meeting. And thanks for posting the article on your blog.
I took your article and asked the genetic genealogist that I consult with on ChatGPT to review the article. I also used the “search” feature to find available info on the public internet to respond to your presentation.
Here is the response: https://chatgpt.com/share/68c7dc60-7420-8003-b547-6ad99139e010 ChatGPT consulted 30+ sources, which you can see. All I checked were live links! Most of the sources referred to in the response are from Roberta Estes, DNA Painter, and gedmatch.
Source: Arthur Sissman – prompting in collaboration with ChatGPT 15 Sep 2025 Remember: AI is your assistant, not your researcher. You are the human in the loop 🙏🏼: Caveat Lector ⛔️ Trust but VERIFY 👍
Regards, Arthur Naples, FL 954-328-3559 genresearch13@yahoo.com
PS: I am now conducting “Clinic” group on how to use Chatbots and Prompt/Context Writing. The clinics are interactive, hopefully the attendee has 2 monitors, etc, and is watching on one and doing on another. I have one Clinic group that decided to get together again! This is all free.
>
LikeLiked by 1 person
Thank you very much for your feedback and effort to get a ChatGPT review. All Segmentologisits should follow that link and see all the feedback. Significant, IMO, is that the feedback is from other genetic genealogists. I’m hoping we can come to some consensus on this topic.
My second ever blogpost was: Benefits of Triangulation on 9 May 2015 and it included 16 benefits, but didn’t explicitly state that a TG based on my Matches was a segment of my DNA! I missed the most important benefit. The corollary is that each of the Matches will have their own TG, which overlaps mine as shown in my TG.
Thanks, again, Jim
LikeLiked by 1 person
My take is that whatever you actually call it, the segment you’ve marked in blue is the one that is significant within your genome. The fact that the inner segment between the blue lines is the only part that triangulates with everyone is far less interesting! So this seems more a discussion of semantics: the larger blue segment is the most useful and meaningful part since it may reveal recombination points in your DNA.
LikeLiked by 1 person
Thanks, Jonny – loved your talk at ECGGC. And agree with your take on the blue TG. When I’m Painting at your site, that’s the important roll-up segment to paint – mine! Jim
LikeLike
Ciao jim.dei segmenti più grandi in una triangolazione ho su mhyeritage ho altre aziende diciamo segmenti triangolati piccoli tra 7,1-9cM con un gruppo di matches tra 10cM o 14 matches che triangola no con segmenti di 7,1-fino ha 9cM che possibilità che rientrano nelle ultime generazioni.insomma possono essere lo tani ho in un arco genealogico?
LikeLike
I am sure you’re first diagram with the longer triangulated segment is correct.
The reason why any match might be shorter is because the other person’s segment had a crossover on the way down from the common ancestor.
The proof of the pudding is that if you compare the Match 1 peso with the Match 2 person, they should both match each other in all their green areas they both share with you, and possibly more if they both share all the way to the left or right of your green triangulated region.
LikeLiked by 1 person
Thanks for your feedback. The longer TG is my DNA segment, but the Matches have their own, overlapping, (TG) segments. In this example, yes all of the pair-wise green areas Triangulate. Jim
LikeLike
This is an interesting question. I prefer the first version that shows the full extent of the matches, not the second, which limits the matches to only the segment that contains all of the matches. The second version is limited, with only the small segment, which could be quite small in terms of cms. In the first version, there is a triangulation within the triangulation, which is considerably larger than the segment in the second version. I would feel more confident with the larger segment in the first version. Also, the first version extends the segment, which could be larger than represented by only my test. Recognizing that the segment could be larger for other tests gives more opportunities to find more matches and their family trees. Quite a few of my 1st-3rd cousins have done DNA testing, so it isn’t unusual for their tests to have longer segments and more matches than mine does. Also, the first version is more open to possibilities, i.e. more matches, more information.
LikeLiked by 1 person