Family Group Sheets
One of the key features of my Common Ancestor Spreadsheet (see post here) is that it offers an arrangement like a traditional genealogy Family Group Sheet (FGS). The FGS has an Ancestor couple at the top of the sheet, with a list of their children down the page with birth, death, marriage dates and places. If we are going to create an inventory of our DNA Matches with known links to an MRCA, this FGS spreadsheet format would be a great way to do that. It also turns out to be a handy tool when working with Pro Tools.
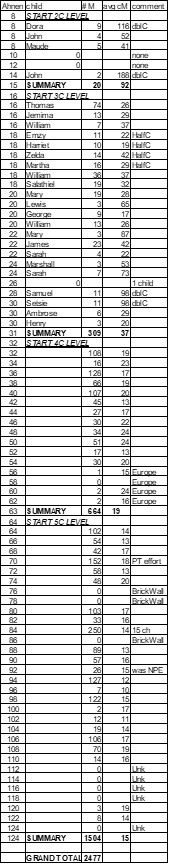
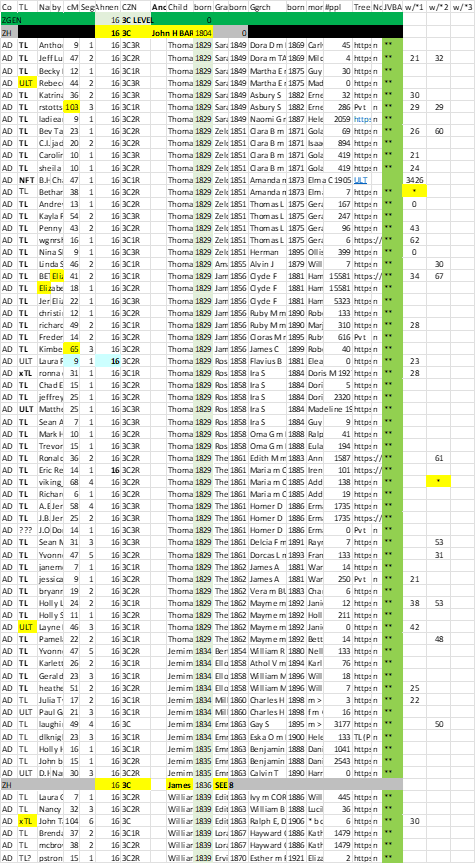
The Common Ancestor spreadsheet for Match cousins is actually a “nested” FGS. By sorting on Ancestor Ahnentafel Numbers, all the Matches connected to one Ancestor are grouped together. By also sorting on the birth year of the Ancestor’s children, this “FGS sort” results with Matches grouped under each child. By adding sorts on birth years for grandchildren and great grandchildren, we get a “nested” FGS. I regularly use my entire spreadsheet sorted by these four columns.
This arrangement has several advantages when using Pro Tools…
1. When Pro Tools indicates a parent/child or sibling relationship to an existing Match (already entered into the spreadsheet), I can create a new row and copy most of the info and just adjust one column – a real time saver. And this works even with new Matches with No Tree, Private Tree, Unlinked Tree, Scrawny Tree, even small cMs – Pro Tools has already provided all the relationship information needed.
2. When Pro Tools indicates a (full) 1C relationship to an existing Match, this limits the relationship possibilities to only two. [In my experience, 1C estimates are highly accurate.] Analysis: the new Match is connected to the existing Match (already in the spreadsheet) on (1) the same side I am on, or (2) on the other side. Be aware of this! If the new Match is on the “other” side, they are NOT part of this Ancestor (Ahnentafel) line. If the new Match has any info in a Tree, this “side” issue can usually be figured out and the spreadsheet cells filled out (mostly by copying from the existing Match). If there is no Tree info, the “side” can usually be determined by looking at the Shared Matches of the new Match (sorted on new Match’s cMs). There should be a clear consensus (at/near the top of that list) of the same Ancestor line as the existing Match. If not, then skip this new Match. If so, I add a row for the new Match, copy data from the existing Match, and enter GUESS for the new Match parent (as a sibling of the existing Match parent), and then the new Match [NB: to save typing, I indicate each “terminal” Match as an asterisk (*) because they are already spelled out in the Match column near the beginning of the row.]
Analysis summary: A) look at their Tree; and/or B) look at their closest SMOMs.
3. For a 1C1R or 1C2R the estimates are still very good, and the process above can be used. Use available info or judgement to shift the new Match to the right or left per the “removes”. Where the individuals are not known, just put Unknown or Private in the cell. The complete path down to the Match is not critical, IMO.
4. When Pro Tools indicate Aunt/Uncle or Niece/Nephew, that too is highly accurate, as are the genders. Similar to the above, there is usually enough information to place them in the spreadsheet (which is like a horizontal Tree).
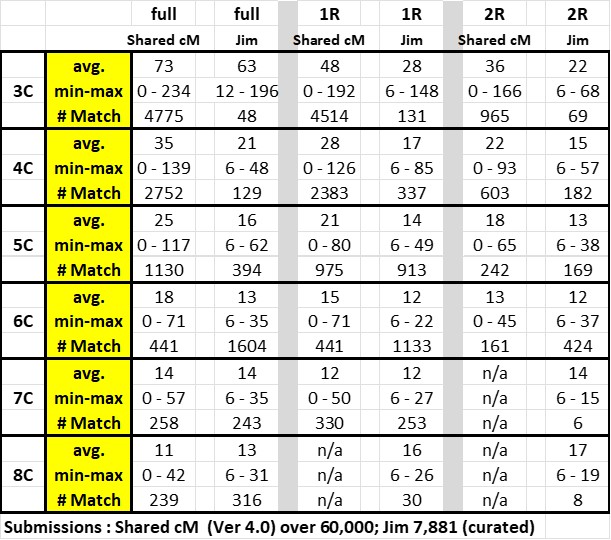
5. Pro Tools often includes a Half relationship in their estimate. This is based on tables that indicate two estimates shown are almost exactly the same cM range. Although technically correct, it is much more likely, IMO, that the relationships are standard (NOT Half). But a few will be Half so watch for that situation. Remember these Pro Tools cMs are between your Match and the Shared Match (not affected by whether or not you have a Half relationships with the Ancestor)
6. Adding a hitherto unknown child branch – best described by a recent example I had. In looking for my A38 (ALLEN ancestor) cousins, I found a bunch descending from four well documented children of A38 – 56 Match cousins (4C, 4C1R, 4C2R and 4C3R) with an average of 18cM. There appears to be more than four children in the 1810-30 Virginia census records. And there was an old story about this family, that a son named William went west. So when some known Matches had some SMOMs with ancestor William H ALLEN born 1815 in VA and living in IL, I took notice – it seemed to fit. As I pushed it with Pro Tools I found (so far) 10 Matches descending from William H ALLEN averaging 20cM. But more importantly, those Matches also had Shared Matches with 12 of the 56 Matches from other children from this A38. It sure looked like a Cluster with gray cells to other Clusters! I’d really like to determine William’s Y-DNA; and/or some DNA segment data… But, in the meantime, I’ve got two of William’s descendants checking their Matches for links to my A38 ALLEN. There are 147 Trees at Ancestry for William H ALLEN – not a one has any good clue to his ancestry, except that he was born in VA. Not my Brick Wall, but I think there will be 147 happy campers.
A key point in this long story, is the DNA has no sense of geography. The facts that four children stayed in VA (and were well known) and one child moved far away, made no difference to the DNA. From each descendant’s viewpoint, all the lines were equal – and a pretty even distribution of Matches showed up for all 5 children. The DNA is like blind justice.
7. Equality – a final thought is that this spreadsheet is a lot like the DNA – it’s relatively equal over all the Ancestors and descendants. This spreadsheet encourages me to treat all of my Ancestors equally (they each have an Ahnentafel placeholder row). I still have my “favorite” Ancestors, but as I methodically go through the spreadsheet, I’m spending time on each one. This includes the Ancestors that have issues… This spreadsheet also highlights the Brick Wall holes, to be plugged with floating family branches. This is a good thing.
To me, the key points in doing this spreadsheet work also include:
1. An inventory of Matches who have MRCAs with me. Separate from my on-line Tree. Saved in the cloud and/or archived – available to my heirs or selected genealogy archives someday.
2. Family Group Sheets – of sorts* – this is a standard genealogy tool.
3. A Quality Control check on the accuracy of name spelling and birth years; and the FGS itself. This QC review often reveals “quirks” (as a kinder word) that folks have in their Trees…
4. With Ancestor second marriages, this FGS listing will show the demarcation between full cousins and Half cousins. [I add “INSIGHT” rows with marriage years that will sort and separate the children to the different parent couples.] Half cousins for me only occur at the children level in my spreadsheet. Half cousins between Matches and Shared Matches can occur anywhere.
5. A re-sort by Match name highlights multiple relationships. Since shared DNA is divided by 4 (on average) going back each generation, the closer relationships are much more likely. I’ve found some Matches with MRCAs on both sides of my Tree. With single shared segments, the DNA can only come from one Ancestor. With multiple shared segments, there may be a segment for each line.
* I used “of sorts” in 2 above, because this FGS will not usually be a complete list of all Ancestor children, grandchildren, etc. It includes only the ones who provided a DNA path down to our Matches. Which in turn depends on family sizes and who did DNA tests – there can be wide variations on both.
Note: If I were starting over, I’d probably add name & birth year columns for 9 generations – out to 8C level; and then a catch-all column for any additional info. This would provide a handy way to evaluate the cousinship levels. Reminder: I only list the given name and one initial for males; and the given name, initial and married surname for females. I try to keep it as easy and simple as possible.
Bottom line: An FGS spreadsheet offers an easy way to add new Matches which have been identified by Pro Tools as closely related to known Matches. This adds independent, genealogy triangulation and tight Clusters to an inventory of known Matches. It will be an outstanding adjunct to an auto-Clustering program.
Also – you don’t have to use a spreadsheet to benefit from most of the concepts imbedded above.
[22CZ] Segment-ology: Pro Tools Part 18 – Family Group Sheets by Jim Bartlett 20241209