A Segment-ology TIDBIT
A question recently came up: Are the Ancestors on two sides of a crossover point, always a mother and father (in either order)? Or: If I know the Common Ancestor (i.e. the father or the mother of the TG couple) of a TG segment, must the next TG segment be the other parent of the TG couple.? The answer is YES, with an important caveat: only when we are talking about mother and father of our Ancestor who created the crossover.
Important scientific fact: A crossover is formed when a human recombines two Chromosomes to create a new Chromosome that is then passed to a child. One of the two Chromosomes is from the Mother, and the other is from the Father. So one parent is on one side of each crossover, and the other parent is on the other side of the crossover. Here is Figure 6 from my 2015 blogpost: Segments – Bottom Up:

Note: each of the Chr 05 lines above is your Maternal Chr 05 – it’s just broken down for each generation. In the Grandparent look, the two crossovers were created by the parent using grandparent segments (assuming an average of 2 crossovers per generation for Chr 05). Note the Ahnentafel numbers to represent generic ancestors – even numbers are males, odd numbers are females. The first crossover created by the parent shows 7 & 6, or female & male, on the two sides of the crossover. When the first grandparent segment ends at the crossover, the next segment is the opposite parent. The second crossover created by the parent has 6 & 7 (male & female) on the two sides of the crossover.
The next line – the Great grandparent look has 2 more crossovers – created by the grandparents, when each of them recombined their respective 2 Great grandparent chromosomes. One of the crossovers is between 14 & 15 and the other between 13 & 12 (there was no crossover when the Ancestor 14 segment was passed to daughter 7). So again, each new crossover has a male and a female (in some order) on the two sides of each crossover.
Check out the two crossovers (on average) added at each of the next two generations – they all have the mother on one side and the father on the other side of the crossover. Note carefully the word “added” (or created or formed).
Now here is the catch… In the Great grandparent look above, the last crossover has 12 & 14 on each side – two males. This seems to contradict the basic concept. And if we were applying the basic concept to TGs at the Great grandparent level it would be wrong. What’s up? Well, what looks like a crossover between Ancestors 12 & 14 is in fact a crossover – but it was formed by Ancestor 3 when she recombined Chr 06s from her parents 6 and 7 – these are the two parents of the ancestor who first formed (or added or created) the crossover.
When we form Triangulated Groups (TGs), we use groups of overlapping segments. But there is nothing in the TG criteria about the generation of the TG. We do understand that the TGs start and end at crossover points – when we shift from one Ancestor’s DNA to another Ancestor’s DNA. But until we can Walk the Segments Back (generation by generation), we don’t know when the crossovers were formed. There is one generation for each crossover, but until we have Chromosome Mapping we don’t know which generation it is.
Note: A TG Summary Spreadsheet will give good clues to the formation of crossover points – see Observation 5 (see linked blogpost). In generation after generation the older crossovers can be seen, with only about 2 new crossovers in each generation. So the farther back we go with Chromosome Mapping, the newly formed crossovers will be there (with mother and father on the two sides). But the other crossovers may not appear to be mother/father, unless the origin of the crossover can be determined.
Bottom Line: With TG segments, sometimes the next TG on a chromosome will be the other parent, but more often it will not.
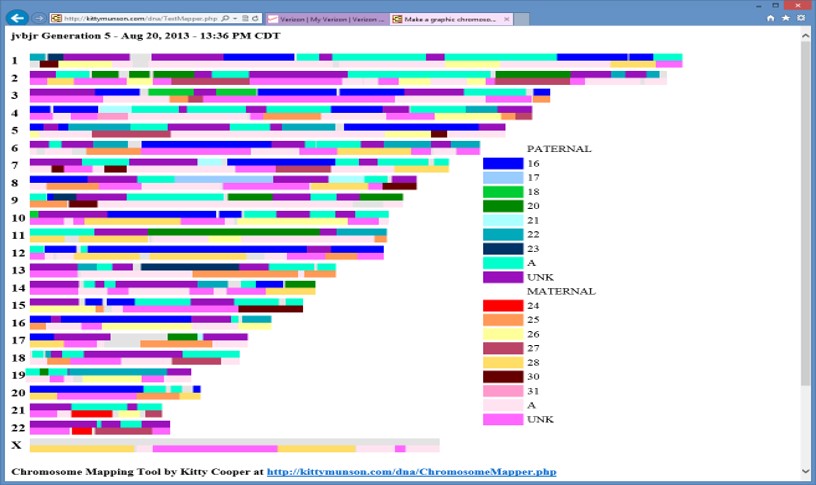
Edit 20240403: It was suggested that I add a Chromosome Map, showing segments from my 16 2xG grandparents. Here is one I did in 2013:
[22CA] Segment-ology: What is the Next Segment? TIDBIT by Jim Bartlett 20231209
